327D
 
 | |
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
2RQE
 
 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3V89
 
 | | The crystal structure of transferrin binding protein A (TbpA) from Neisseria meningitidis serogroup B in complex with the C-lobe of human transferrin | | Descriptor: | Serotransferrin, Transferrin-binding protein A | | Authors: | Noinaj, N, Oke, M, Easley, N, Zak, O, Aisen, P, Buchanan, S.K. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
2MFF
 
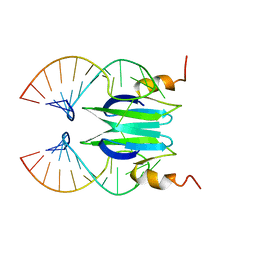 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
1OL1
 
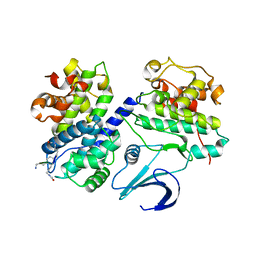 | | Cyclin A binding groove inhibitor H-Cit-Cit-Leu-Ile-(p-F-Phe)-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CIR-CIR-LEU-ILE-PFF-NH2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-08-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
2ME6
 
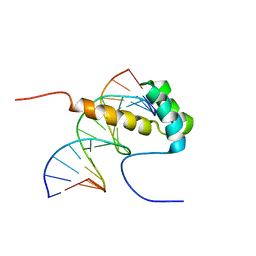 | | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*TP*AP*AP*TP*TP*AP*GP*TP*CP*G)-3'), Homeobox protein GBX-1 | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1 from Homo sapiens in complex with the DNA sequence CGACTAATTAGTCG
To be Published
|
|
3A03
 
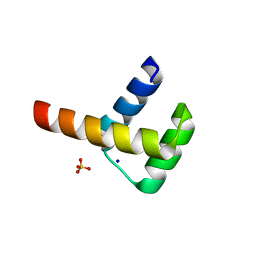 | | Crystal structure of Hox11L1 homeodomain | | Descriptor: | SODIUM ION, SULFATE ION, T-cell leukemia homeobox protein 2 | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
2M1O
 
 | | ID3 stem | | Descriptor: | RNA (5'-R(P*AP*GP*CP*AP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*UP*GP*UP*A)-3') | | Authors: | Popovic, M, Greenbaum, N.L. | | Deposit date: | 2012-12-03 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ID3 stem
To be Published
|
|
3VCB
 
 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3ACD
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with IMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3A29
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2MLL
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
3A4A
 
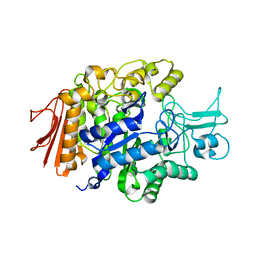 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
3VDF
 
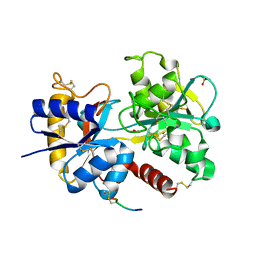 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with diaminopimelic acid at 1.46 A Resolution | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with diaminopimelic acid at 1.46 A Resolution
To be Published
|
|
2ZYF
 
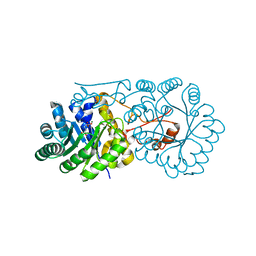 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
3VG4
 
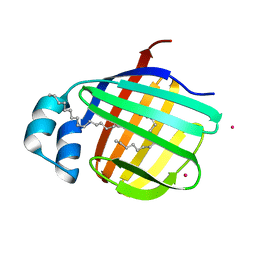 | | Cadmium derivative of human LFABP | | Descriptor: | CADMIUM ION, Fatty acid-binding protein, liver, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
2MJ9
 
 | | Designed Exendin-4 analogues | | Descriptor: | Exendin-4 | | Authors: | Rovo, P, Farkas, V, Straner, P, Szabo, M, Jermendy, A, Hegyi, O, Toth, G.K, Perczel, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-06-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational design of alpha-helix-stabilized exendin-4 analogues.
Biochemistry, 53, 2014
|
|
2ZYJ
 
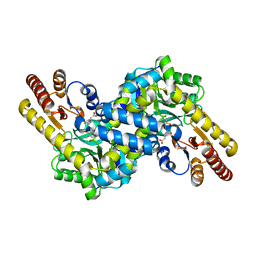 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-glutamate), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Ouchi, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-26 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual roles of a conserved pair, Arg23 and Ser20, in recognition of multiple substrates in alpha-aminoadipate aminotransferase from Thermus thermophilus.
Biochem.Biophys.Res.Commun., 388, 2009
|
|
3VHS
 
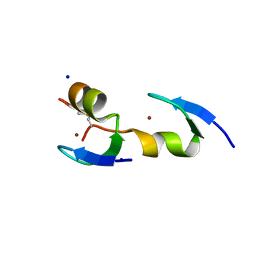 | |
3VI4
 
 | | Crystal structure of alpha5beta1 integrin headpiece in complex with RGD peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of alpha5beta1 integrin ectodomain: Atomic details of the fibronectin receptor
J.Cell Biol., 197, 2012
|
|
3VJR
 
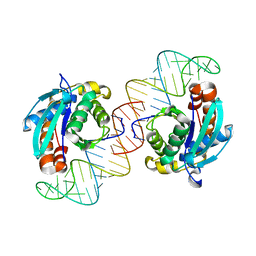 | | Crystal structure of Peptidyl-tRNA hydrolase from Escherichia coli in complex with the CCA-acceptor-T[PSI]C domain of tRNA | | Descriptor: | Peptidyl-tRNA hydrolase, tRNA CCA-acceptor | | Authors: | Ito, K, Murakami, R, Mochizuki, M, Qi, H, Shimizu, Y, Miura, K.I, Ueda, T, Uchiumi, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition and catalysis of peptidyl-tRNA hydrolase.
Nucleic Acids Res., 40, 2012
|
|
322D
 
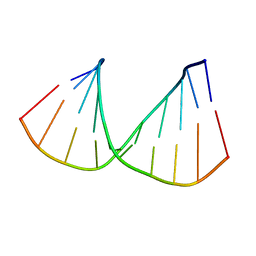 | | CRYSTAL STRUCTURES OF D(CCGGGCCM5CGG)-HEXAGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*CP*CP*(5CM)P*GP*G)-3') | | Authors: | Tippin, D.B, Sundaralingam, M. | | Deposit date: | 1997-03-17 | | Release date: | 1997-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA.
J.Mol.Biol., 267, 1997
|
|
2M51
 
 | |
