4WZX
 
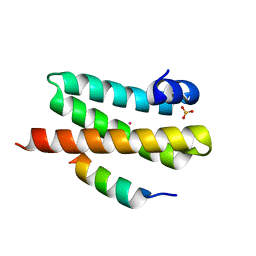 | | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins | | Descriptor: | COBALT (II) ION, IST1 homolog, SULFATE ION, ... | | Authors: | Caballe, A, Wenzel, D.M, Agromayor, M, Alam, S.L, Skalicky, J.J, Kloc, M, Carlton, J.G, Labrador, L, Sundquist, W.I, Martin-Serrano, J. | | Deposit date: | 2014-11-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3859 Å) | | Cite: | ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Elife, 4, 2015
|
|
8UH6
 
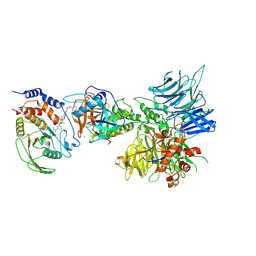 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
4X8O
 
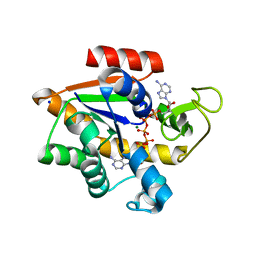 | | Crystal structure of E. coli Adenylate kinase Y171W mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4TTY
 
 | |
4TOY
 
 | |
4X6I
 
 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | Descriptor: | 2-amino-4-bromo-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}benzamide, Cathepsin K, SULFATE ION | | Authors: | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
4X3Z
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
4X28
 
 | | Crystal structure of the ChsE4-ChsE5 complex from Mycobacterium tuberculosis | | Descriptor: | Acyl-CoA dehydrogenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Unraveling Cholesterol Catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha 2 beta 2 Acyl-CoA Dehydrogenase Initiates beta-Oxidation of 3-Oxo-cholest-4-en-26-oyl CoA.
Acs Infect Dis., 1, 2015
|
|
4X8M
 
 | | Crystal structure of E. coli Adenylate kinase Y171W mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
4RO2
 
 | | Crystal Structure of CNG mimicking NaK-ETPP mutant cocrystallized with Methylammonium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1BFD
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
4XCP
 
 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XOU
 
 | | Crystal structure of the SR Ca2+-ATPase in the Ca2-E1-MgAMPPCP form determined by serial femtosecond crystallography using an X-ray free-electron laser. | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Nass, K, Drachmann, N.D, Markvardsen, A.J, Gutmann, M.J, Barends, T.R.M, Mattle, D, Shoeman, R.L, Doak, R.B, Boutet, S, Messerschmidt, M, Seibert, M.M, Williams, G.J, Foucar, L, Reinhard, L, Sitsel, O, Gregersen, J.L, Clausen, J.D, Boesen, T, Gotfryd, K, Wang, K.-T, Olesen, C, Moller, J.V, Nissen, P, Schlichting, I. | | Deposit date: | 2015-01-16 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser.
Iucrj, 2, 2015
|
|
4XJI
 
 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TNL
 
 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4XJD
 
 | | X-ray structure of Lysozyme2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJH
 
 | | X-ray structure of LysozymeS1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TNC
 
 | |
4XD7
 
 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
4XUW
 
 | | Structure of the hazelnut allergen, Cor a 8 | | Descriptor: | Non-specific lipid-transfer protein, PHOSPHATE ION | | Authors: | Offermann, L.R, Perdue, M.L, Bublin, M, Pfeifer, S, Dubiela, P, Hoffmann-Sommergruber, K, Chruszcz, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.
J.Agric.Food Chem., 63, 2015
|
|
