4EK7
 
 | | High speed X-ray analysis of plant enzymes at room temperature | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
4EGD
 
 | | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom crystal structure of native hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
2LD2
 
 | |
2PE0
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOINOSITIDE-DEPENDENT PROTEIN KINASE 1 (PDK1) 5-Hydroxy-3-[1-(1H-pyrrol-2-yl)-eth-(Z)-ylidene]-1,3-dihydro-indol-2-one COMPLEX | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-HYDROXY-3-[(1R)-1-(1H-PYRROL-2-YL)ETHYL]-2H-INDOL-2-ONE, GLYCEROL, ... | | Authors: | Whitlow, M, Adler, M. | | Deposit date: | 2007-04-01 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Indolinone based phosphoinositide-dependent kinase-1 (PDK1) inhibitors. Part 1: Design, synthesis and biological activity.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3WKG
 
 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
2LJ5
 
 | |
2LNH
 
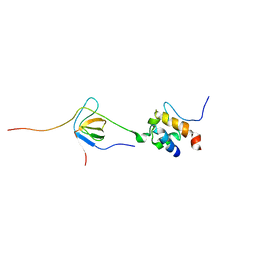 | | Enterohaemorrhagic E. coli (EHEC) exploits a tryptophan switch to hijack host F-actin assembly | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, Neural Wiskott-Aldrich syndrome protein, Secreted effector protein EspF(U) | | Authors: | Aitio, O, Hellman, M, Skehan, B, Kesti, T, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2011-12-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Structure, 20, 2012
|
|
2LEG
 
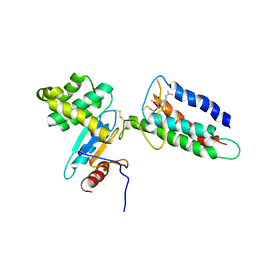 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
4XB9
 
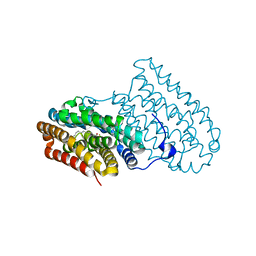 | |
1LRP
 
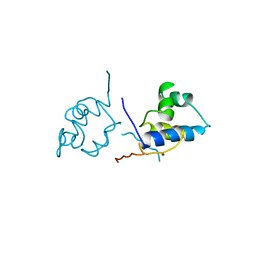 | |
3WMP
 
 | | Crystal structure of SLL-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WN1
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
2LPU
 
 | | Solution structures of KmAtg10 | | Descriptor: | KmAtg10 | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
3WC1
 
 | | Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a G-1 deleted tRNA(His) | | Descriptor: | 75-mer tRNA, Likely histidyl tRNA-specific guanylyltransferase | | Authors: | Nakamura, A, Nemoto, T, Sonoda, T, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Structural basis of reverse nucleotide polymerization
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LCM
 
 | | NMR structure of S3-4 peptide | | Descriptor: | Voltage-dependent N-type calcium channel subunit alpha-1B | | Authors: | Douzi, B, Darbon, H, De waar, M. | | Deposit date: | 2011-05-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | structure of S4-3 voltage sensor peptide
To be Published
|
|
2LLG
 
 | | NMR structure of the protein NP_814968.1 from Enterococcus faecalis | | Descriptor: | Uncharacterized protein | | Authors: | Susac, L, Serrano, P, Geralt, M, Mohanty, B, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-11-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_814968.1 from Enterococcus faecalis
To be Published
|
|
2OM7
 
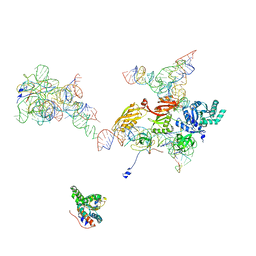 | | Structural Basis for Interaction of the Ribosome with the Switch Regions of GTP-bound Elongation Factors | | Descriptor: | 16S ribosomal RNA (H5), 30S ribosomal protein S12, 30S ribosomal protein S2, ... | | Authors: | Connell, S.R, Wilson, D.N, Rost, M, Schueler, M, Giesebrecht, J, Dabrowski, M, Mielke, T, Fucini, P, Spahn, C.M.T. | | Deposit date: | 2007-01-21 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors.
Mol.Cell, 25, 2007
|
|
3WP3
 
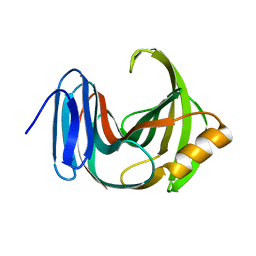 | |
7ZQK
 
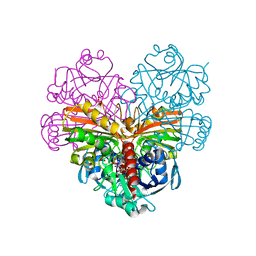 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
2LOR
 
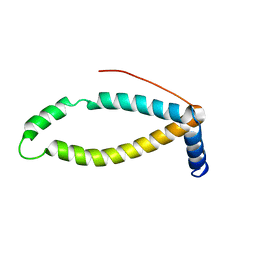 | | Backbone structure of human membrane protein TMEM141 | | Descriptor: | Transmembrane protein 141 | | Authors: | Bayrhuber, M, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
7ZQ4
 
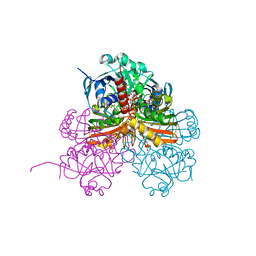 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
2LFF
 
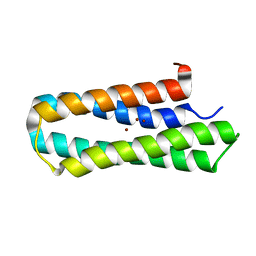 | | Solution structure of Diiron protein in presence of 8 eq Zn2+, Northeast Structural Genomics consortium target OR21 | | Descriptor: | Diiron protein, ZINC ION | | Authors: | Pires, M, Wu, Y, Mills, J.L, Reig, A, Englander, W, Degrado, W, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Diiron protein in presence of 8 eq Zn2+
To be Published
|
|
8A4T
 
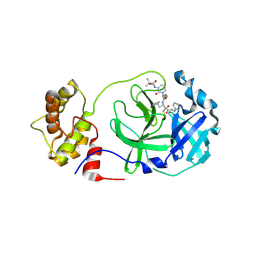 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
3WFI
 
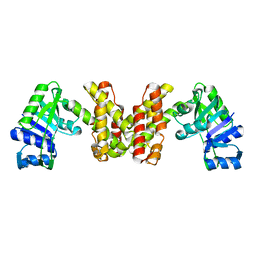 | | The crystal structure of D-mandelate dehydrogenase | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
2LGC
 
 | |
