4OTD
 
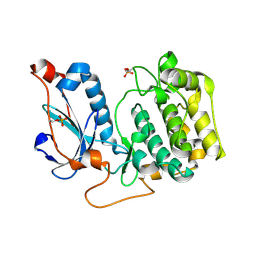 | | Crystal Structure of PRK1 Catalytic Domain | | Descriptor: | Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
5H46
 
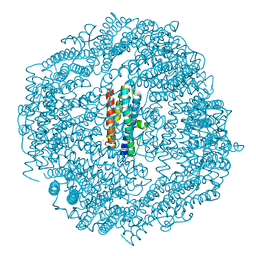 | | Mycobacterium smegmatis Dps1 mutant - F47E | | Descriptor: | DNA protection during starvation protein, FE (II) ION | | Authors: | Williams, S.M, Chandran, A.V, Vijayan, M, Chatterji, D. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage.
Structure, 25, 2017
|
|
7TPS
 
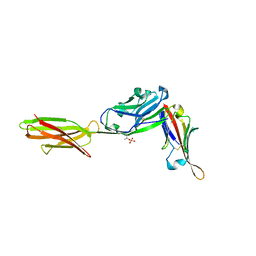 | | Crystal structure of ALPN-202 (engineered CD80 vIgD) in complex with PD-L1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demonte, D.W, Maurer, M.F, Akutsu, M, Kimbung, Y.R, Logan, D.T, Walse, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 costimulation for anti-tumor immunity.
Nat Commun, 13, 2022
|
|
3BE4
 
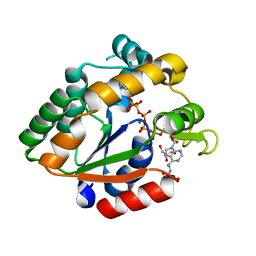 | | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, GLYCEROL, ... | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Cryptosporidium parvum adenylate kinase cgd5_3360.
To be Published
|
|
4LA6
 
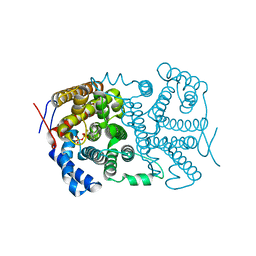 | |
5G4G
 
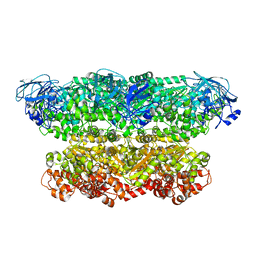 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G64
 
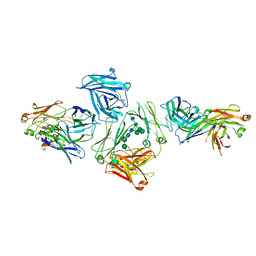 | | The complex between human IgE-Fc and two anti-IgE Fab fragments | | Descriptor: | FAB FRAGMENT, IG EPSILON CHAIN C REGION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Davies, A.M, Allan, E.G, Keeble, A.H, Delgado, J, Cossins, B.P, Mitropoulou, A.N, Pang, M.O.Y, Ceska, T, Beavil, A.J, Craggs, G, Westwood, M, Henry, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.715 Å) | | Cite: | Allosteric mechanism of action of the therapeutic anti-IgE antibody omalizumab.
J. Biol. Chem., 292, 2017
|
|
5G2V
 
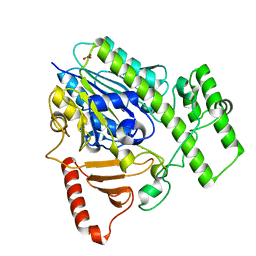 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3BFT
 
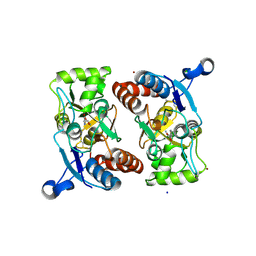 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
4OY7
 
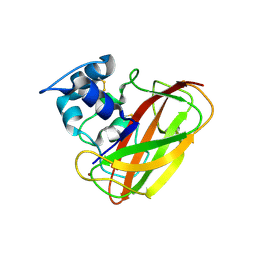 | | Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Putative secreted cellulose binding protein | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5G36
 
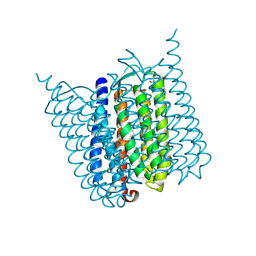 | | Yellow form of Halorhodopsin from Halobacterium salinarum in a new rhombohedral crystal form | | Descriptor: | CHLORIDE ION, HALORHODOPSIN, RETINAL, ... | | Authors: | Schreiner, M, Schlesinger, R, Heberle, J, Niemann, H.H. | | Deposit date: | 2016-04-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Halobacterium Salinarum Halorhodopsin with Partially Depopulated Primary Chloride Binding Site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7TT7
 
 | | BamABCDE bound to substrate EspP in the barrelized EspP/continuous open BamA state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
7TJ8
 
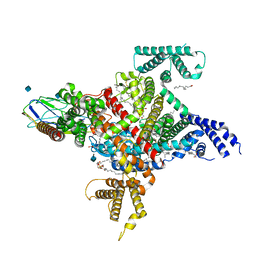 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in nanodiscs | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
5G4F
 
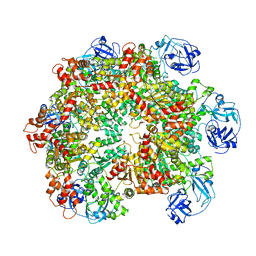 | | Structure of the ADP-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3BH1
 
 | | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, UPF0371 protein DIP2346 | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Gilmore, M, Iizuka, M, Groshong, C, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae.
To be Published
|
|
7TT6
 
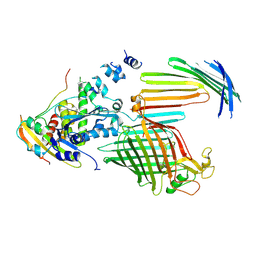 | | BamABCDE bound to substrate EspP in the intermediate-open EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
5GAH
 
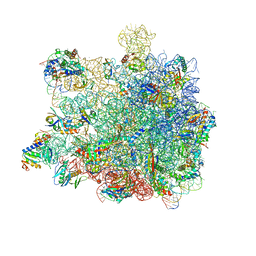 | | RNC in complex with SRP with detached NG domain | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
5I6H
 
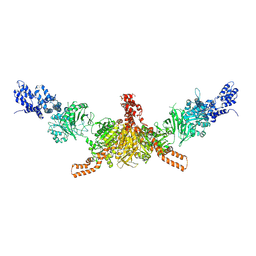 | | Crystal structure of CD-CT domains of Chaetomium thermophilum acetyl-CoA carboxylase | | Descriptor: | Acetyl-CoA carboxylase-like protein | | Authors: | Hunkeler, M, Stuttfeld, E, Hagmann, A, Imseng, S, Maier, T. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (7.2 Å) | | Cite: | The dynamic organization of fungal acetyl-CoA carboxylase.
Nat Commun, 7, 2016
|
|
2PEL
 
 | | PEANUT LECTIN | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEANUT LECTIN, ... | | Authors: | Banerjee, R, Das, K, Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation, protein-carbohydrate interactions and a novel subunit association in the refined structure of peanut lectin-lactose complex.
J.Mol.Biol., 259, 1996
|
|
3WJZ
 
 | | Orotidine 5'-monophosphate decarboxylase D75N mutant from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujihashi, M, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
7TT3
 
 | | BamABCDE bound to substrate EspP class 5 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT4
 
 | | BamABCDE bound to substrate EspP class 6 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
3BJP
 
 | | Urate oxidase cyanide uric acid ternary complex | | Descriptor: | ACETYL GROUP, CYANIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Prange, T, Colloc'h, N, El Hajji, M, Castro, B, Chiadmi, M. | | Deposit date: | 2007-12-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: Mechanistic implications
Bmc Struct.Biol., 8, 2008
|
|
7TSZ
 
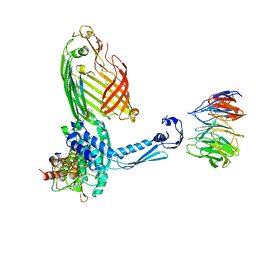 | | BamABCDE bound to substrate EspP class 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
