7NLP
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-canavanine | | Descriptor: | Acetylglutamate kinase, L-CANAVANINE, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNC
 
 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal-5'-phosphate and 6-Methoxyquinoline-3-carboxylic acid | | Descriptor: | 6-methoxyquinoline-3-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLX
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 7-(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 7-(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNI
 
 | |
7NLW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(5-methoxy-1H-indol-3-yl)acetonitrile | | Descriptor: | 2-(5-methoxy-1~{H}-indol-3-yl)ethanenitrile, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in apo form. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6SQL
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and N-(3-(aminomethyl)phenyl)-5-chloro-3-methylbenzo[b]thiophene-2-sulfonamide | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-[3-(aminomethyl)phenyl]-5-chloranyl-3-methyl-1-benzothiophene-2-sulfonamide | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
3KVU
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA FlK - T42S mutant in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Fluoroacetyl-CoA thioesterase FlK | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
1NK1
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
6SQ5
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid | | Descriptor: | (~{E})-3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
5FPK
 
 | | MONOMERIC RADA IN COMPLEX WITH FATA TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTG PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
6SQ7
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-(4-chloro-3-nitrobenzoyl)benzoic acid | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1K9Y
 
 | | The PAPase Hal2p complexed with magnesium ions and reaction products: AMP and inorganic phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, Halotolerance protein HAL2, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
3KUV
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - T42S mutant in complex with acetate. | | Descriptor: | ACETATE ION, Fluoroacetyl coenzyme A thioesterase | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KX8
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK | | Descriptor: | fluoroacetyl-CoA thioesterase | | Authors: | Chirgadze, D.Y, Dias, M.V.B, Huang, F, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3KV8
 
 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK - Wild type FlK in complex with fluoro-acetate | | Descriptor: | Fluoroacetyl-CoA thioesterase FlK, fluoroacetic acid | | Authors: | Dias, M.V.B, Huang, F, Chirgadze, D.Y, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-11-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3N76
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with compound 5 | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
1SMR
 
 | |
1JP4
 
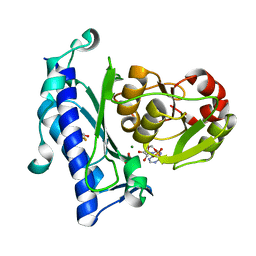 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
3TM7
 
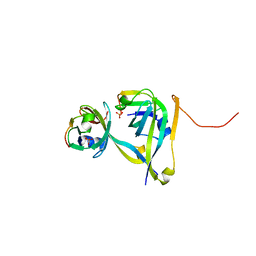 | | Processed Aspartate Decarboxylase Mutant with Asn72 mutated to Ala | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Webb, M.E, Lobley, C.M.C, Soliman, F, Kilkenny, M.L, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2011-08-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Escherichia coli aspartate alpha-decarboxylase Asn72Ala: probing the role of Asn72 in pyruvoyl cofactor formation
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3COW
 
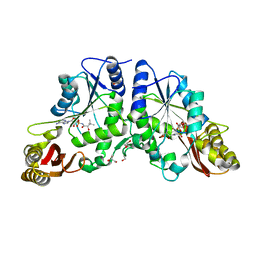 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution- in complex with sulphonamide inhibitor 2 | | Descriptor: | 5'-O-{[(2R)-2-hydroxy-3,3-dimethylbutanoyl]sulfamoyl}adenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2008-03-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate.
Chembiochem, 9, 2008
|
|
