5EZG
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 22 at 1.84A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(1-pyrimidin-2-ylpiperidin-4-yl)methyl]pyrrolidine-1-carboxamide | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F0H
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 28 at 1.99A resolution | | Descriptor: | 3-[1-(4-cyanophenyl)piperidin-4-yl]-~{N}-[(4-piperidin-1-ylphenyl)methyl]propanamide, HTH-type transcriptional regulator EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F1J
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 1 at 1.63A resolution | | Descriptor: | 3-cyclopentyl-1-pyrrolidin-1-yl-propan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F0C
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium tuberculosis in complex with compound 4 at 1.87A resolution | | Descriptor: | 3-cyclopentyl-~{N}-(3-piperidin-1-ylphenyl)propanamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5EZH
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 21 at 1.7A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(1-pyridin-2-ylpiperidin-4-yl)methyl]pyrrolidine-1-carboxamide | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F0F
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 15 at 1.76A resolution | | Descriptor: | 4-[4-(3-oxidanylidene-3-pyrrolidin-1-yl-propyl)piperidin-1-yl]benzenecarbonitrile, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F27
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 2 at 1.68A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, ~{N}-methyl-1-(4-piperidin-1-ylphenyl)methanamine | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5F04
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 3 at 1.84A resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl-1-[3-[4-(methylaminomethyl)phenyl]-1,3-diazinan-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
4GCR
 
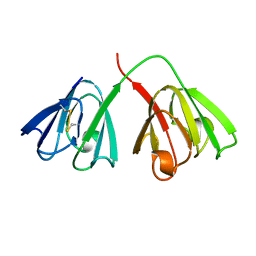 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4A6X
 
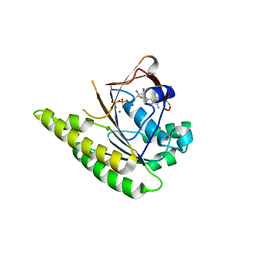 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4A6P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4MUN
 
 | |
3KGV
 
 | |
7YY0
 
 | |
7YY5
 
 | |
7YYB
 
 | |
7YY6
 
 | |
7YY1
 
 | |
7YY9
 
 | |
7YY3
 
 | |
7YYC
 
 | |
7YWM
 
 | |
7YY2
 
 | |
7YY7
 
 | |
7YY8
 
 | |
