2JGD
 
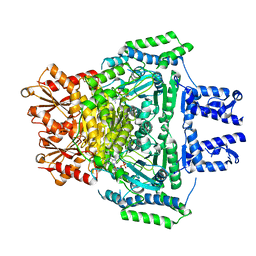 | | E. COLI 2-oxoglutarate dehydrogenase (E1o) | | Descriptor: | 2-OXOGLUTARATE DEHYDROGENASE E1 COMPONENT, ADENOSINE MONOPHOSPHATE | | Authors: | Frank, R.A.W, Price, A.J, Northrop, F.D, Perham, R.N, Luisi, B.F. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
J.Mol.Biol., 368, 2007
|
|
2FIC
 
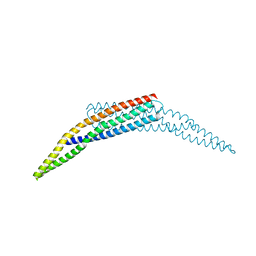 | | The crystal structure of the BAR domain from human Bin1/Amphiphysin II and its implications for molecular recognition | | Descriptor: | Myc box-dependent-interacting protein 1, XENON | | Authors: | Casal, E, Federici, L, Zhang, W, Fernandez-Recio, J, Priego, E.M, Miguel, R.N, Duhadaway, J.B, Prendergast, G.C, Luisi, B.F, Laue, E.D. | | Deposit date: | 2005-12-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crystal Structure of the BAR Domain from Human Bin1/Amphiphysin II and Its Implications for Molecular Recognition
Biochemistry, 45, 2006
|
|
4CDI
 
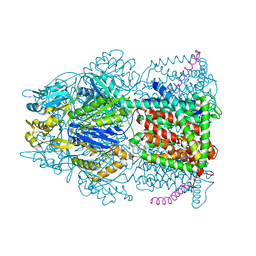 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, PREDICTED PROTEIN | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the Acrab-Tolc Multidrug Efflux Pump.
Nature, 509, 2014
|
|
4C48
 
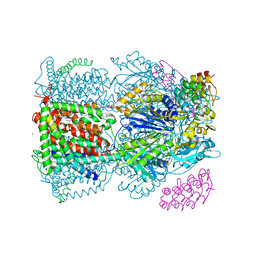 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-09-02 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the AcrAB-TolC multidrug efflux pump.
Nature, 509, 2014
|
|
4AM3
 
 | | Crystal structure of C. crescentus PNPase bound to RNA | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AIM
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
1YC9
 
 | | The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 resolution | | Descriptor: | MERCURY (II) ION, multidrug resistance protein, octyl beta-D-glucopyranoside | | Authors: | Federici, L, Du, D, Walas, F, Matsumura, H, Fernandez-Recio, J, McKeegan, K.S, Borges-Walmsley, M.I, Luisi, B.F, Walmsley, A.R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the outer membrane protein VCEC from the bacterial pathogen vibrio cholerae at 1.8 A resolution
J.Biol.Chem., 280, 2005
|
|
4AID
 
 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4ATO
 
 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | Authors: | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2012-05-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8BVM
 
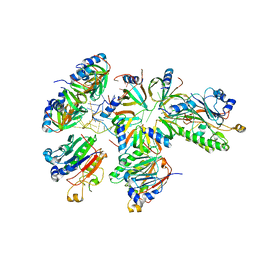 | |
8BVJ
 
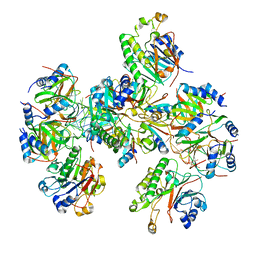 | | Hfq-Crc-estA translation repression complex | | Descriptor: | Catabolite repression control protein, RNA-binding protein Hfq, estA mRNA | | Authors: | Dendooven, T, Luisi, B.F. | | Deposit date: | 2022-12-04 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding.
Embo J., 42, 2023
|
|
8BVH
 
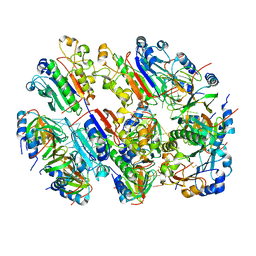 | |
8CGL
 
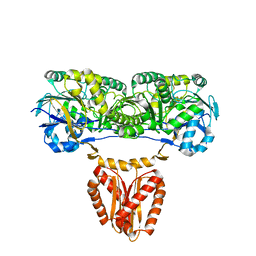 | |
6SGR
 
 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc with cardiolipin | | Descriptor: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
6SGU
 
 | | Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
6SGT
 
 | | Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc with cardiolipin | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
2C0B
 
 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C4R
 
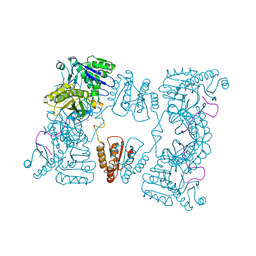 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, SSRNA MOLECULE: 5'-R(*AP*CP*AP*GP*UP*AP*UP*UP*UP*GP)-3', ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Luisi, B.F. | | Deposit date: | 2005-10-21 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2CE1
 
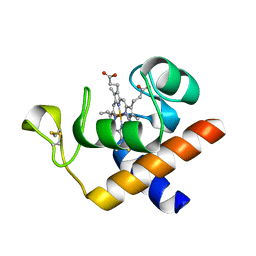 | | Structure of reduced Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-02 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
2CE0
 
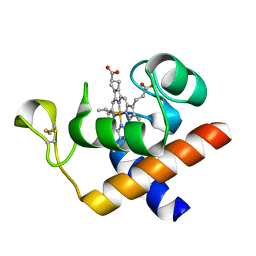 | | Structure of oxidized Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-01 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
2BP7
 
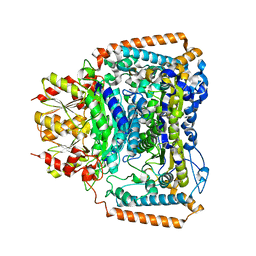 | | New crystal form of the Pseudomonas putida branched-chain dehydrogenase (E1) | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2005-04-18 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Structure, 13, 2005
|
|
2BX2
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, RNA (5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP* UP*GP*UP*U)-3'), ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
6SGS
 
 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc | | Descriptor: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
2C8M
 
 | |
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
