6DCX
 
 | | iASPP-PP-1c structure and targeting of p53 | | Descriptor: | RelA-associated inhibitor, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Glover, J.N.M, Zhou, Y, Edwards, R.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Structure, 27, 2019
|
|
3D8A
 
 | | Co-crystal structure of TraM-TraD complex. | | Descriptor: | Protein traD, Relaxosome protein TraM | | Authors: | Glover, J.N.M, Lu, J, Wong, J.J, Edwards, R.A. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation.
Mol.Microbiol., 70, 2008
|
|
5U6K
 
 | |
1FOS
 
 | | TWO HUMAN C-FOS:C-JUN:DNA COMPLEXES | | Descriptor: | C-JUN PROTO-ONCOGENE PROTEIN, DNA (5'-D(*AP*AP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*AP*T)-3'), ... | | Authors: | Glover, J.N.M, Harrison, S.C. | | Deposit date: | 1995-03-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA.
Nature, 373, 1995
|
|
1QVP
 
 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
1QW1
 
 | | Solution Structure of the C-Terminal Domain of DtxR residues 110-226 | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6Y7F
 
 | | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7) | | Descriptor: | CHLORIDE ION, Elongation of very long chain fatty acids protein 7, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Nie, L, Pike, A.C.W, Bushell, S.R, Chu, A, Cole, V, Speedman, D, Rodstrom, K.E.J, Kupinska, K, Shrestha, L, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of human ELOVL fatty acid elongase 7 (ELOVL7)
TO BE PUBLISHED
|
|
4MEV
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNC
 
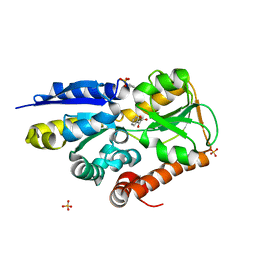 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNI
 
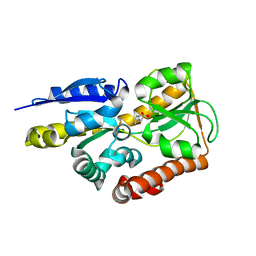 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | Descriptor: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5HFK
 
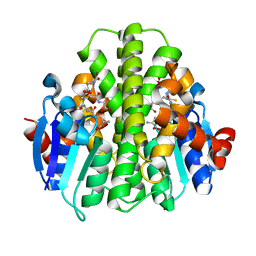 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
4LYY
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677.
To be Published
|
|
4MCI
 
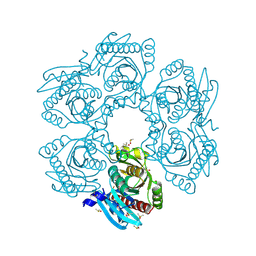 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520. | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520.
To be Published
|
|
4MF5
 
 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
6R7X
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-10, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Baronina, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R7Z
 
 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4MP4
 
 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4MK3
 
 | | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h) | | Descriptor: | Glutathione S-transferase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, MAGNESIUM ION, ... | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
To be published
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
6R7Y
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6R65
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4B8N
 
 | | Cytochrome b5 of Ostreococcus tauri virus 2 | | Descriptor: | CYTOCHROME B5-HOST ORIGIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isupov, M, Reid, E.L, Weynberg, K.D, Love, J, Wilson, W.H, Kelly, S.L, Lamb, D.C, Allen, M.J, Littlechild, J.A. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterisation of a Viral Cytochrome B5.
FEBS Lett., 587, 2013
|
|
