3UWB
 
 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
3UWA
 
 | | Crystal structure of a probable peptide deformylase from synechococcus phage S-SSM7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RIIA-RIIB membrane-associated protein, ZINC ION | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
4DR8
 
 | | Crystal structure of a peptide deformylase from Synechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Lorimer, D, Abendroth, J, Craig, T, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
4DR9
 
 | | Crystal structure of a peptide deformylase from synechococcus elongatus in complex with actinonin | | Descriptor: | ACTINONIN, BROMIDE ION, Peptide deformylase, ... | | Authors: | Lorimer, D, Abendroth, J, Craig, T, Burgin, A, Segall, A, Rohwler, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
5V8T
 
 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | Descriptor: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
8DOF
 
 | | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044 | | Descriptor: | (2R)-2-cyclohexyl-2-[(4-{[5-(propan-2-yl)-1H-pyrazol-3-yl]amino}-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]ethan-1-ol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Horanyi, P.S, Wang, Y, Todd, M.H, Abendroth, J, Edwards, T, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044
To Be Published
|
|
1XMJ
 
 | | Crystal structure of human deltaF508 human NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
8DSR
 
 | | Structure of Plasmepsin X (PM10, PMX) from Plasmodium falciparum 3D7 in complex with UCB7362 | | Descriptor: | (2E,6S)-6-{2-chloro-3-[(2-cyclopropylpyrimidin-5-yl)amino]phenyl}-2-imino-6-methyl-3-[(2S,4S)-2-methyloxan-4-yl]-1,3-diazinan-4-one, Plasmepsin X | | Authors: | Abendroth, J, Lorimer, D.D. | | Deposit date: | 2022-07-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Characterization of Potent, Efficacious, Orally Available Antimalarial Plasmepsin X Inhibitors and Preclinical Safety Assessment of UCB7362 .
J.Med.Chem., 65, 2022
|
|
1XMI
 
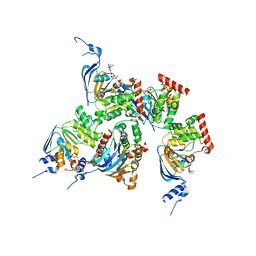 | | Crystal structure of human F508A NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
9BR4
 
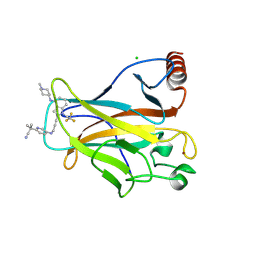 | | Crystal structure of p53 Y220C mutant in complex with PC-9859 | | Descriptor: | 2-methyl-2-{5-[(3-{4-[(1-methylpiperidin-4-yl)amino]-1-(2,2,2-trifluoroethyl)-1H-indol-2-yl}prop-2-yn-1-yl)amino]pyridin-2-yl}propanenitrile, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Abendroth, J, Lorimer, D.D, Vu, B, Takana, N. | | Deposit date: | 2024-05-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Rezatapopt (PC14586), a First-in-Class, Small-Molecule Reactivator of p53 Y220C Mutant in Development.
Acs Med.Chem.Lett., 16, 2025
|
|
9BR3
 
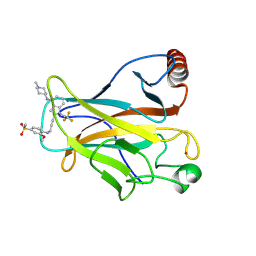 | | Crystal structure of p53 Y220C mutant in complex with PC-10709 | | Descriptor: | 2-{3-[4-(methanesulfonyl)-2-methoxyanilino]prop-1-yn-1-yl}-N-(1-methylpiperidin-4-yl)-1-(2,2,2-trifluoroethyl)-1H-indol-4-amine, Cellular tumor antigen p53, ZINC ION | | Authors: | Abendroth, J, Lorimer, D.D, Vu, B, Tanaka, N. | | Deposit date: | 2024-05-10 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restoration of the Tumor Suppressor Function of Y220C-Mutant p53 by Rezatapopt, a Small Molecule Reactivator.
Cancer Discov, 2025
|
|
8DGD
 
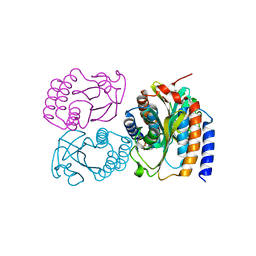 | | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GAF domain-containing protein, ZINC ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Delker, S.L, Weiss, M.J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-06-23 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of GAF domain-containing protein, from Klebsiella pneumoniae
to be published
|
|
4NIM
 
 | |
4NOZ
 
 | | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Organic hydroperoxide resistance protein | | Authors: | Dranow, D.M, Lukacs, C.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia
TO BE PUBLISHED
|
|
4TWR
 
 | | Structure of UDP-glucose 4-epimerase from Brucella abortus | | Descriptor: | NAD binding site:NAD-dependent epimerase/dehydratase:UDP-glucose 4-epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of UDP-glucose 4-epimerase from Brucella melitensis
To Be Published
|
|
3VFI
 
 | | Crystal Structure of a Metagenomic Thioredoxin | | Descriptor: | thioredoxin | | Authors: | Craig, T.K, Gardberg, A, Lorimer, D.D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Metagenomic Thioredoxin
To be Published
|
|
6U94
 
 | | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344 | | Descriptor: | GLYCEROL, RND efflux system, outer membrane lipoprotein, ... | | Authors: | Horanyi, P.S, Fox III, D, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-09-06 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344
To Be Published
|
|
1Q2W
 
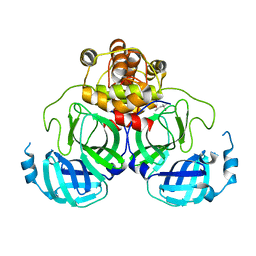 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
5K85
 
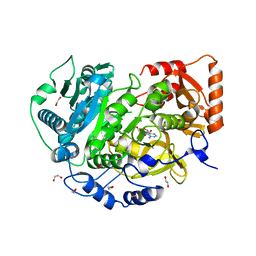 | | Crystal Structure of Acetyl-CoA Synthetase in Complex with Adenosine-5'-propylphosphate and Coenzyme A from Cryptococcus neoformans H99 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Delker, S.L, Potts, K.T, Lorimer, D.D, Edwards, T.E, Mutz, M.W, SSGCID | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ACETYL-COA SYNTHETASE IN COMPLEX WITH ADENOSINE-5'-PROPYLPHOSPHATE AND COENZYME A FROM CRYPTOCOCCUS NEOFORMANS H99
To Be Published
|
|
5K8F
 
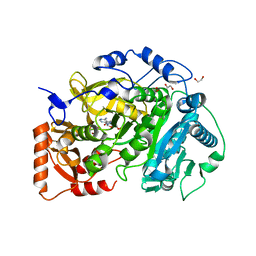 | | Crystal structure of Acetyl-CoA Synthetase in complex with ATP and Acetyl-AMP from Cryptococcus neoformans H99 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Delker, S.L, Potts, K.T, Lorimer, D.D, Edwards, T.E, Mutz, M.W. | | Deposit date: | 2016-05-30 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Acetyl-CoA Synthetase in complex with ATP and Acetyl-AMP from Cryptococcus neoformans H99
To Be Published
|
|
5DLC
 
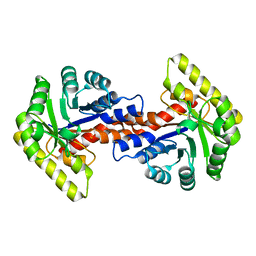 | |
4NPS
 
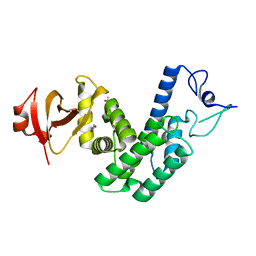 | | Crystal Structure of Bep1 protein (VirB-translocated Bartonella effector protein) from Bartonella clarridgeiae | | Descriptor: | ACETATE ION, Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Microorganisms, 9, 2021
|
|
4XIN
 
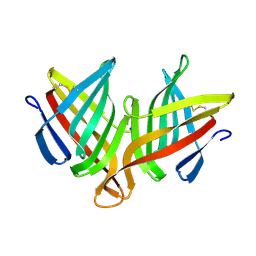 | |
6X9F
 
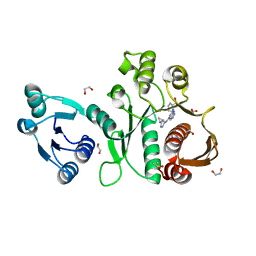 | | Pseudomonas aeruginosa MurC with AZ8074 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ8074
To Be Published
|
|
6X9N
 
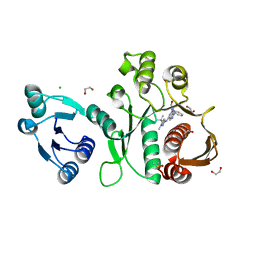 | | Pseudomonas aeruginosa MurC with AZ5595 | | Descriptor: | (2R)-2-({4-[(5-tert-butyl-1-methyl-1H-pyrazol-3-yl)amino]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}amino)-2-phenylethan-1-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ5595
To Be Published
|
|
