7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3TJ1
 
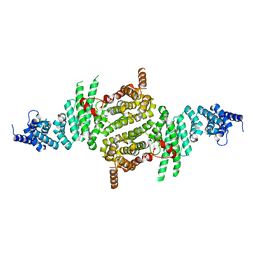 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3NFI
 
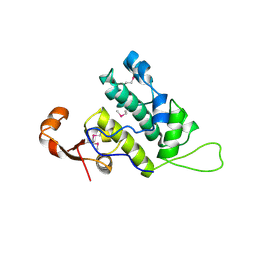 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
8QFG
 
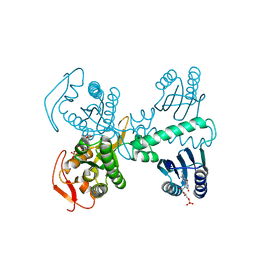 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC after 5 seconds of blue light illumination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFE
 
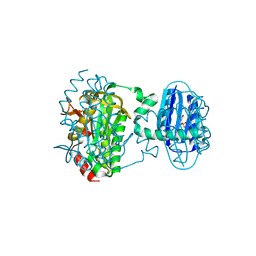 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, MAGNESIUM ION | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFF
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFH
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFJ
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 2.3 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFI
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 1.8 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Sato, T, Round, A, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
3NFF
 
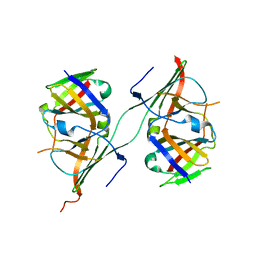 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | Descriptor: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3NFH
 
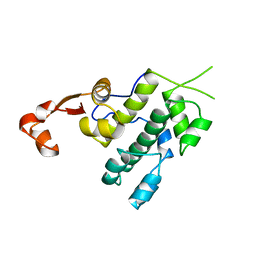 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 (P4) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
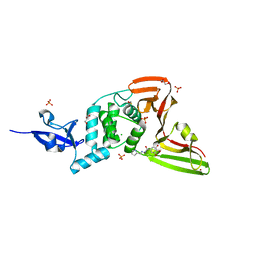 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7QCK
 
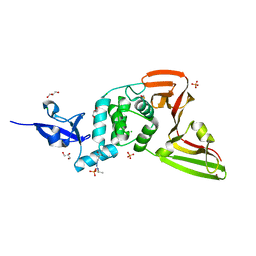 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
