1O4B
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU83876. | | Descriptor: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-3,4-DIPHOSPHONOPHENYLALANINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O42
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU81843. | | Descriptor: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-O-PHOSPHONOTYROSINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4E
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78299. | | Descriptor: | 2,6-DIFORMYL-4-METHYLPHENYL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4R
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78783. | | Descriptor: | (PHENYL-PHOSPHONO-METHYL)-PHOSPHONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O44
 
 | | Crystal structure of sh2 in complex with ru85052 | | Descriptor: | 2-{4-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-CARBOXY-PHENYL}-MALONIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4C
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHOSPHATE. | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4I
 
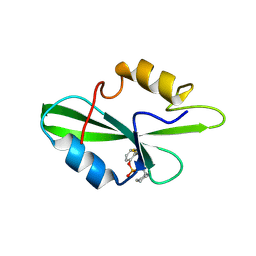 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PAS219. | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, cyclohexylmethyl 2-formylphenyl hydrogen (S)-phosphate | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O48
 
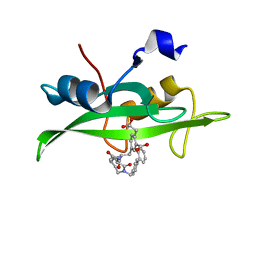 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85053. | | Descriptor: | 5-[2-ACETYLAMINO-2-(1-BIPHENYL-4-YLMETHYL-2-OXO-AZEPAN-3-YLCARBAMOYL)-ETHYL]-2-CARBOXYMETHYL-BENZOIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4O
 
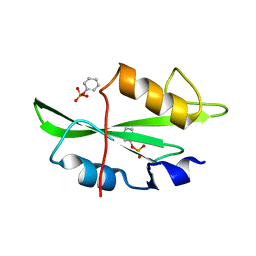 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHENYLPHOSPHATE. | | Descriptor: | PHENYL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4Q
 
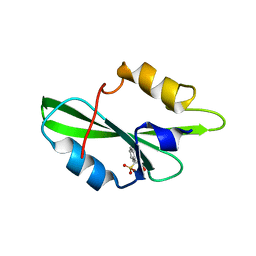 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79256. | | Descriptor: | PHENYL(SULFO)ACETIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4N
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH OXALIC ACID. | | Descriptor: | OXALIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4D
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78262. | | Descriptor: | 2-FORMYLPHENYL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
5LRG
 
 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin B complex | | Descriptor: | Anabaenopeptin B, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
3LFS
 
 | | Crystal structure of CDK2 with SAR37, an aminoindazole type inhibitor | | Descriptor: | Cell division protein kinase 2, N-(6-chloro-5-phenyl-1H-indazol-3-yl)butanamide | | Authors: | Dreyer, M.K, Wendt, K.U, Schimanski-Breves, S, Loenze, P. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of potent GSK3beta inhibitors with selectivity for Cdk1 and Cdk2.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LFQ
 
 | | Crystal structure of CDK2 with SAR60, an aminoindazole type inhibitor | | Descriptor: | Cell division protein kinase 2, N-(6,7-difluoro-5-phenyl-1H-indazol-3-yl)butanamide | | Authors: | Dreyer, M.K, Wendt, K.U, Schimanski-Breves, S, Loenze, P. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of potent GSK3beta inhibitors with selectivity for Cdk1 and Cdk2.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LFN
 
 | | Crystal structure of CDK2 with SAR57, an aminoindazole type inhibitor | | Descriptor: | Cell division protein kinase 2, N-[6-(4-hydroxyphenyl)-5-phenyl-1H-indazol-3-yl]butanamide | | Authors: | Dreyer, M.K, Wendt, K.U, Schimanski-Breves, S, Loenze, P. | | Deposit date: | 2010-01-18 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Rational design of potent GSK3beta inhibitors with selectivity for Cdk1 and Cdk2.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5LRK
 
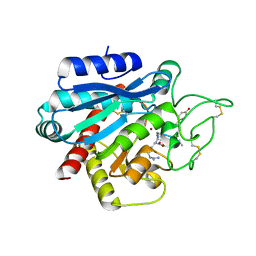 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin F complex | | Descriptor: | Anabaenopeptin F, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
5LRJ
 
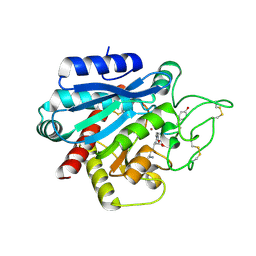 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin C complex | | Descriptor: | Anabaenopeptin C, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
5CGJ
 
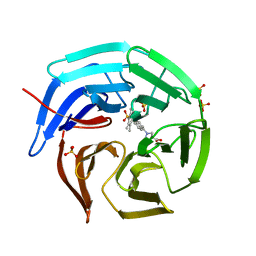 | | Crystal structure of murine Keap1 in complex with RA839, a non-covalent small-molecule binder to Keap1 and selective activator of Nrf2 signalling. | | Descriptor: | (3S)-1-(4-{[(2,3,5,6-tetramethylphenyl)sulfonyl]amino}naphthalen-1-yl)pyrrolidine-3-carboxylic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Schimanski-Breves, S, Loenze, P, Engel, C.K. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Characterization of RA839, a Noncovalent Small Molecule Binder to Keap1 and Selective Activator of Nrf2 Signaling.
J.Biol.Chem., 290, 2015
|
|
2BMG
 
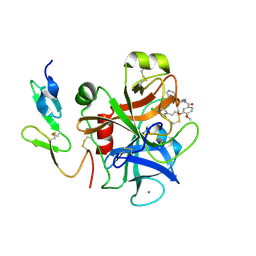 | | Crystal structure of factor Xa in complex with 50 | | Descriptor: | 3-[2-(2,4-DICHLOROPHENYL)ETHOXY]-4-METHOXY-N-[(1-PYRIDIN-4-YLPIPERIDIN-4-YL)METHYL]BENZAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Schreuder, H, Matter, H, Will, D.W, Nazare, M, Laux, V, Wehner, V, Loenze, P, Liesum, A. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Requirements for Factor Xa Inhibition by 3-Oxybenzamides with Neutral P1 Substituents: Combining X-Ray Crystallography, 3D-Qsar and Tailored Scoring Functions
J.Med.Chem., 48, 2005
|
|
5LYF
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{R})-1-[[(1~{R},2~{S},4~{S})-2-bicyclo[2.2.1]heptanyl]amino]-3-cyclohexyl-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
5LYD
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-(sulfamoylamino)hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
5LYL
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-2-[[(2~{S})-1-(1-adamantylamino)-3-cyclohexyl-1-oxidanylidene-propan-2-yl]sulfamoylamino]-6-azanyl-hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
5LYI
 
 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(1~{R},2~{S},4~{R})-1,7,7-trimethyl-2-bicyclo[2.2.1]heptanyl]amino]propan-2-yl]sulfamoylamino]hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
