3J7N
 
 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7L
 
 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
6VVO
 
 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
8EDX
 
 | |
5GPO
 
 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
2QZ9
 
 | | crystal structure of aspartate semialdehyde dehydrogenase II from vibrio cholerae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Viola, R.E, Liu, X, Ohren, J.F, Faehnle, C.R. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a redundant enzyme: a second isoform of aspartate beta-semialdehyde dehydrogenase in Vibrio cholerae.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3L7W
 
 | |
3L8R
 
 | | The crystal structure of PtcA from S. mutans | | Descriptor: | Putative PTS system, cellobiose-specific IIA component | | Authors: | Lei, J, Liu, X, Li, L. | | Deposit date: | 2010-01-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PtcA from Streptococcus mutans
To be Published
|
|
6LJB
 
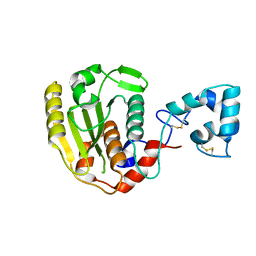 | |
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
7TBI
 
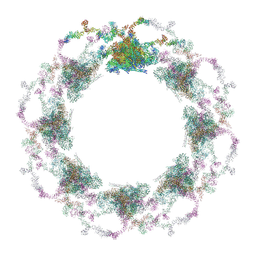 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | Descriptor: | Dyn2, Nic96 R1, Nic96 R2, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
3NR2
 
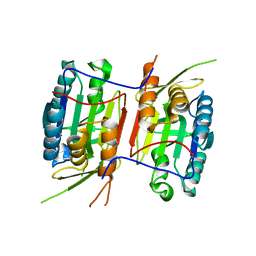 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3CXS
 
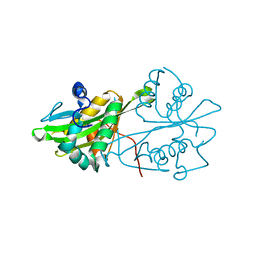 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
7F05
 
 | |
3CXQ
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
2VAS
 
 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
3OD5
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
2VB6
 
 | | Myosin VI (MD-insert2-CaM, Delta Insert1) Post-rigor state (crystal form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
3L9T
 
 | |
3LAS
 
 | | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution | | Descriptor: | GLYCEROL, GUANIDINE, MAGNESIUM ION, ... | | Authors: | Ma, L.-L, Wang, K.-T, Liu, X, Su, X.-D. | | Deposit date: | 2010-01-07 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution
To be published
|
|
6L31
 
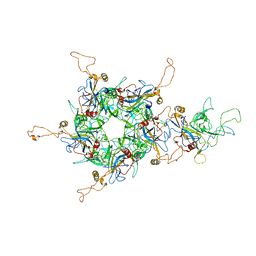 | | L1 protein of human papillomavirus 6 | | Descriptor: | Major capsid protein L1 | | Authors: | Li, S.W, Liu, X.L, Gu, Y. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection.
Emerg Microbes Infect, 8, 2019
|
|
3L7X
 
 | |
3L87
 
 | |
