5HC6
 
 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in apo form | | Descriptor: | SULFATE ION, prenyltransference for protein | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6WQA
 
 | | 2.0A angstrom A2a adenosine receptor structure using XFEL data collected in helium atmosphere. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Lee, M.-Y, Geiger, J, Ishchenko, A, Han, G.W, Barty, A, White, T.A, Gati, C, Batyuk, A, Hunter, M.S, Aquila, A, Boutet, S, Weierstall, U, Cherezov, V, Liu, W. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing the power of an X-ray laser for serial crystallography of membrane proteins crystallized in lipidic cubic phase
Iucrj, 7, 2020
|
|
5HC7
 
 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in complex with S-thiolo-isopentenyldiphosphate | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, prenyltransference for protein | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6PNY
 
 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
6M24
 
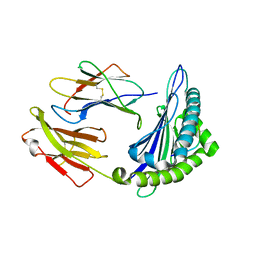 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-2 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2J
 
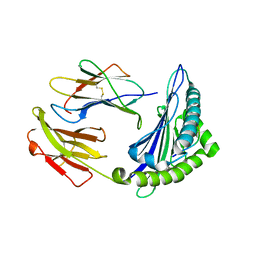 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2K
 
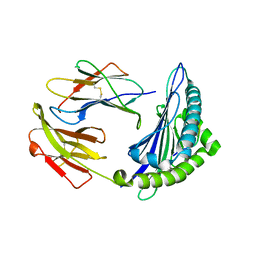 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
8JG4
 
 | |
6PQ0
 
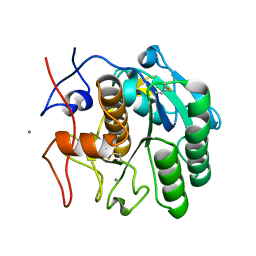 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
6PQ4
 
 | | LCP-embedded Proteinase K treated with lipase | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
2YHV
 
 | |
4ANL
 
 | |
7TD0
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD3
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD2
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD1
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA, state a | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD4
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
8S9Z
 
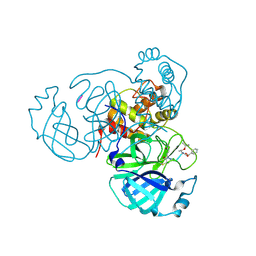 | | Mpro inhibitors of SARS-CoV-2 | | Descriptor: | 3C-like proteinase nsp5, Mpro inhibitor | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of MPI89 with Mpro of SARS-CoV-2 at 1.85A resolution.
To Be Published
|
|
8STY
 
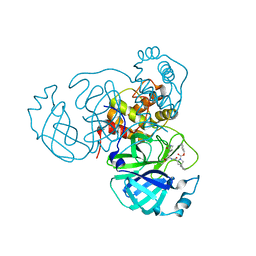 | |
8STZ
 
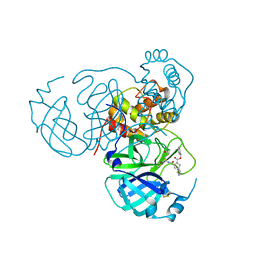 | |
8AZS
 
 | | Type I amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
4ANQ
 
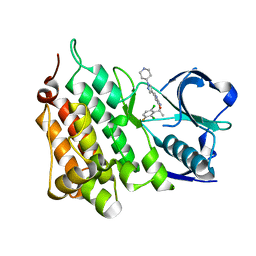 | | Structure of G1269A Mutant Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A. | | Deposit date: | 2012-03-21 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design of Potent and Selective Inhibitors to Overcome Clinical Anaplastic Lymphoma Kinase Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
2YFX
 
 | | Structure of L1196M Mutant Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A. | | Deposit date: | 2011-04-08 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Potent and Selective Inhibitors to Overcome Clinical Anaplastic Lymphoma Kinase Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
3R0H
 
 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
4DJH
 
 | | Structure of the human kappa opioid receptor in complex with JDTic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CITRIC ACID, ... | | Authors: | Wu, H, Wacker, D, Katritch, V, Mileni, M, Han, G.W, Vardy, E, Liu, W, Thompson, A.A, Huang, X.P, Carroll, F.I, Mascarella, S.W, Westkaemper, R.B, Mosier, P.D, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | tructure of the human kappa-opioid receptor in complex with JDTic
Nature, 485, 2012
|
|
