8IF6
 
 | |
5JCE
 
 | | Crystal structure of OsCEBiP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
7QPT
 
 | | Botulinum neurotoxin A4 cell binding domain in complex with GD1a oligosaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type A | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mojanaga, O.O. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
7QPU
 
 | | Botulinum neurotoxin A5 cell binding domain in complex with GM1b oligosaccharide | | Descriptor: | Botulinum neurotoxin sub-type A5, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin Subtypes A4 and A5 Cell Binding Domains in Complex with Receptor Ganglioside.
Toxins, 14, 2022
|
|
6F0P
 
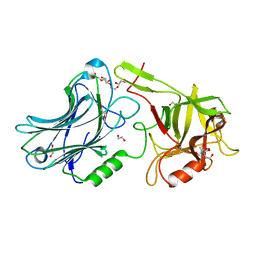 | | Botulinum neurotoxin A4 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, J.R, Rees, J, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
6F0O
 
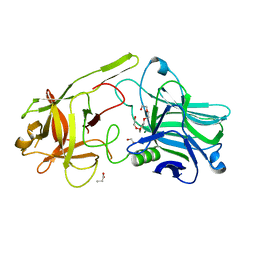 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
6G5G
 
 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6G5K
 
 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B in complex with human synaptotagmin 1 | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-1 | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6G5F
 
 | | Crystal structure of an engineered Botulinum Neurotoxin type B mutant E1191M/S1199Y in complex with human synaptotagmin 1 | | Descriptor: | Botulinum neurotoxin type B, GLYCEROL, MALONATE ION, ... | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6THY
 
 | | Botulinum neurotoxin A3 Hc domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BoNT/A3, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2019-11-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of botulinum neurotoxin subtype A3 cell binding domain in complex with GD1a co-receptor ganglioside.
Febs Open Bio, 10, 2020
|
|
5JCD
 
 | | Crystal structure of OsCEBiP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
4B4A
 
 | | Structure of the TatC core of the twin arginine protein translocation system | | Descriptor: | Lauryl Maltose Neopentyl Glycol, SEC-INDEPENDENT PROTEIN TRANSLOCASE PROTEIN TATC | | Authors: | Rollauer, S.E, Tarry, M.J, Jaaskelainen, M, Graham, J.E, Jaeger, F, Krehenbrink, M, Roversi, P, McDowell, M.A, Stansfeld, P.J, Johnson, S, Liu, S.M, Lukey, M.J, Marcoux, J, Robinson, C.V, Sansom, M.S, Palmer, T, Hogbom, M, Berks, B.C, Lea, S.M. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-05 | | Last modified: | 2012-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Tatc Core of the Twin-Arginine Protein Transport System.
Nature, 49, 2012
|
|
8OW8
 
 | | Crystal Structure of the Catalytic Domain of a Botulinum Neurotoxin Homologue from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, Botulinum-like toxin eBoNT/J light chain, PHOSPHATE ION, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Catalytic Domain of a Botulinum Neurotoxin Homologue from Enterococcus faecium : Potential Insights into Substrate Recognition.
Int J Mol Sci, 24, 2023
|
|
2BKU
 
 | | Kap95p:RanGTP complex | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN RAN, GUANOSINE-5'-TRIPHOSPHATE, IMPORTIN BETA-1 SUBUNIT, ... | | Authors: | Lee, S.J, Matsuura, Y, Liu, S.M, Stewart, M. | | Deposit date: | 2005-02-21 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Nuclear Import Complex Dissociation by Rangtp
Nature, 435, 2005
|
|
7Z5S
 
 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain in complex with GD1a | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin, HEXAETHYLENE GLYCOL, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
7Z5T
 
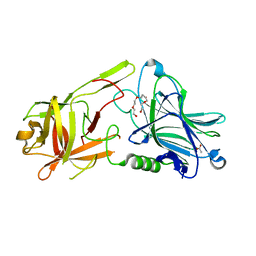 | | Crystal Structure of botulinum neurotoxin A2 cell binding domain | | Descriptor: | ACETATE ION, Botulinum neurotoxin, PENTAETHYLENE GLYCOL | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Mahadeva, T.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Features of Clostridium botulinum Neurotoxin Subtype A2 Cell Binding Domain.
Toxins, 14, 2022
|
|
8AGK
 
 | | Botulinum neurotoxin subtype A6 cell binding domain in complex with GD1a ganglioside | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M, Newell, A.R, Mojanaga, O.O. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8ALP
 
 | | Botulinum neurotoxin A6 cell binding domain crystal form II | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-08-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
8IFG
 
 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
