7C3C
 
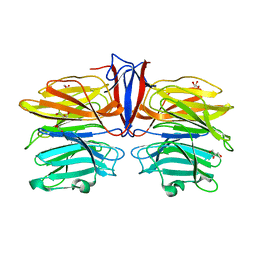 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-manose | | Descriptor: | AofleA, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C37
 
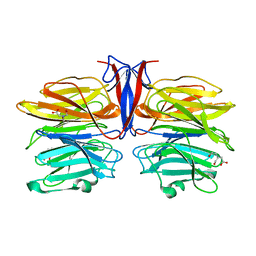 | | Crystal structure of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, BICINE | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
5H7V
 
 | |
4H86
 
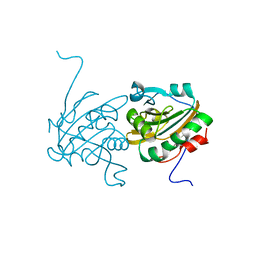 | | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Liu, M, Wang, F, Qiu, R, Wu, T, Gu, S, Tang, R, Ji, C. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form
To be Published
|
|
8K5J
 
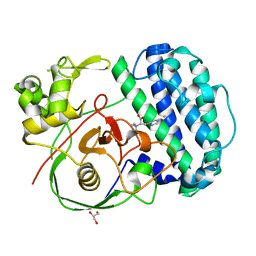 | | The structure of SenA in complex with N,N,N-trimethyl-histidine | | Descriptor: | FE (III) ION, GLYCEROL, N,N,N-trimethyl-histidine, ... | | Authors: | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
8K5K
 
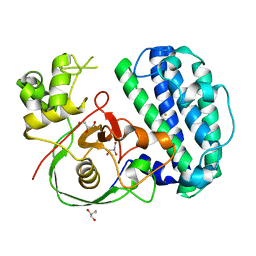 | | The structure of SenA | | Descriptor: | FE (III) ION, GLYCEROL, selenoneine synthase SenA | | Authors: | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
8K5I
 
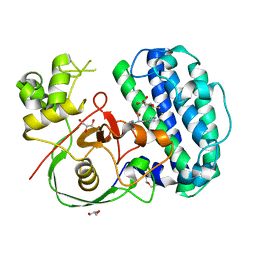 | | The structure of SenA in complex with N,N,N-trimethyl-histidine and thioglucose | | Descriptor: | 1-thio-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
3GKY
 
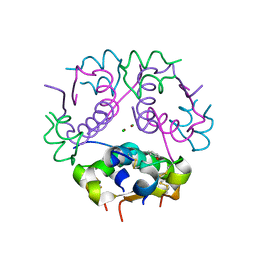 | | The Structural Basis of an ER Stress-Associated Bottleneck in a Protein Folding Landscape | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Liu, M, Wan, Z.L, Chu, Y.C, Alddin, H, Klaproth, B, Weiss, M.A. | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a "nonfoldable" insulin: IMPAIRED FOLDING EFFICIENCY DESPITE NATIVE ACTIVITY.
J.Biol.Chem., 284, 2009
|
|
2LMC
 
 | |
6KY4
 
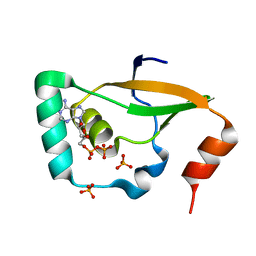 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
7Q34
 
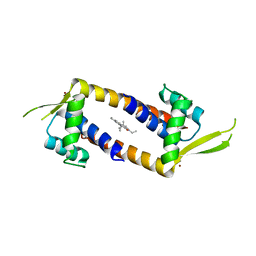 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex squaraine dye | | Descriptor: | 2,4-bis[(E)-(1-ethyl-3,3-dimethyl-indol-2-ylidene)methyl]cyclobutane-1,3-dione, Helix-turn-helix transcriptional regulator, NICKEL (II) ION | | Authors: | Liutkus, M, Mejias, S.H, Barolo, C, Cortajarena, A.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designing Artificial Fluorescent Proteins: Squaraine-LmrR Biophosphors for High Performance Deep-Red Biohybrid Light-Emitting Diodes
Adv Funct Mater, 32, 2022
|
|
8CMQ
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Tb | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CKR
 
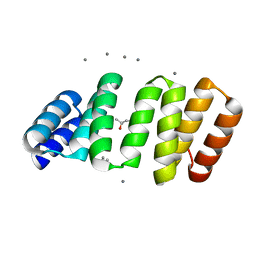 | | Crystal structure of an 8-repeat consensus TPR superhelix with in Hepes with Ca | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Consensus tetratricopeptide repeat protein | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CP8
 
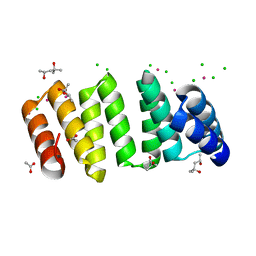 | | Crystal structure of an 8-repeat consensus TPR superhelix with Lead | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CQQ
 
 | |
8CQP
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Calcium (low concentration) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8OT7
 
 | |
8BU0
 
 | | Crystal structure of an 8 repeat consensus TPR superhelix with calcium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CHY
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Zinc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CH0
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix with Gadolinium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Consensus tetratricopeptide repeat protein, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
8CIG
 
 | | Crystal structure of an 8-repeat consensus TPR superhelix in tris Buffer with Calcium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Liutkus, M, Rojas, A.L, Cortajarena, A.L. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse crystalline protein scaffolds through metal-dependent polymorphism.
Protein Sci., 33, 2024
|
|
5HIM
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and dimethylglycine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, N,N-DIMETHYLGLYCINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
7S4V
 
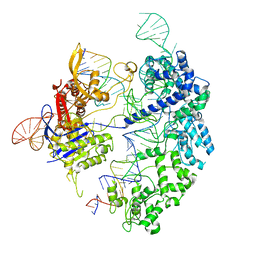 | | Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4U
 
 | | Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand, Target strand, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4X
 
 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
