8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | 分子名称: | Chitin deacetylase, ZINC ION | | 著者 | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | 登録日 | 2022-11-10 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | 著者 | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-05-11 | | 公開日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
7X6S
 
 | |
7X6V
 
 | |
5YLO
 
 | | Structural of Pseudomonas aeruginosa PA4980 | | 分子名称: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | 著者 | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | 登録日 | 2017-10-18 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
5WQE
 
 | |
5WTJ
 
 | | Crystal structure of an endonuclease | | 分子名称: | CRISPR-associated endoribonuclease C2c2 | | 著者 | Liu, L, Wang, Y. | | 登録日 | 2016-12-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
5WTK
 
 | | Crystal structure of RNP complex | | 分子名称: | CRISPR-associated endoribonuclease C2c2, RNA (58-MER) | | 著者 | Liu, L, Wang, Y. | | 登録日 | 2016-12-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
5WNO
 
 | | Crystal structure of C. elegans LET-23 kinase domain complexed with AMP-PNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase let-23 | | 著者 | Liu, L, Thaker, T.M, Jura, N. | | 登録日 | 2017-08-01 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.386 Å) | | 主引用文献 | Regulation of Kinase Activity in the Caenorhabditis elegans EGF Receptor, LET-23.
Structure, 26, 2018
|
|
6IFO
 
 | |
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | 分子名称: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J, Zhu, R, Pei, Y. | | 登録日 | 2021-03-23 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-04-06 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-03-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-06-05 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7E2U
 
 | | Synechocystis GUN4 in complex with phytochrome | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | 著者 | Liu, L, Hu, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
 | | Synechocystis GUN4 in complex with phycocyanobilin | | 分子名称: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | 著者 | Liu, L, Hu, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2R
 
 | |
7E2S
 
 | |
7FH6
 
 | | Friedel-Crafts alkylation enzyme CylK | | 分子名称: | CALCIUM ION, CHLORIDE ION, CylK, ... | | 著者 | Liu, L, Wang, H.Q, Xiang, Z. | | 登録日 | 2021-07-29 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
1M7Q
 
 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | 分子名称: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | 著者 | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | 登録日 | 2002-07-22 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4YVQ
 
 | |
4ZDS
 
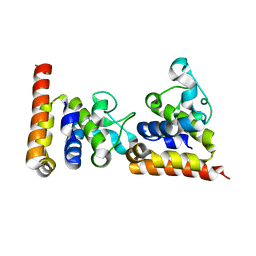 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | 分子名称: | Protein ETHYLENE INSENSITIVE 3 | | 著者 | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | 登録日 | 2015-04-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
6WPG
 
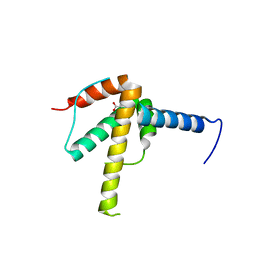 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | 分子名称: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | 著者 | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | 登録日 | 2020-04-27 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.283 Å) | | 主引用文献 | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
5GOH
 
 | | Lys33-linked di-ubiquitin | | 分子名称: | D-ubiquitin, Ubiquitin | | 著者 | Gao, S, Pan, M, Zheng, Y, Liu, L. | | 登録日 | 2016-07-27 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.946 Å) | | 主引用文献 | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOJ
 
 | | Lys63-linked di-ubiquitin | | 分子名称: | D-ubiquitin, Ubiquitin | | 著者 | Gao, S, Pan, M, Zheng, Y, Liu, L. | | 登録日 | 2016-07-27 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
