7RAY
 
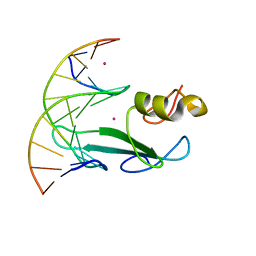 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*A)-R(P*(5MC))-D(P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
8ZMY
 
 | | F0502B-bound WT polymorph 5a alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLO
 
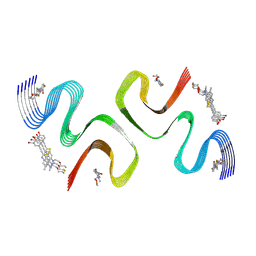 | | F0502B-bound E46K alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLP
 
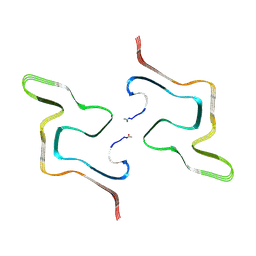 | |
8ZLI
 
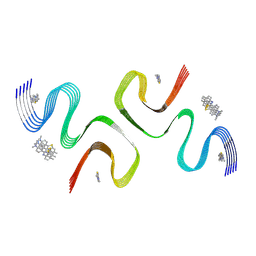 | | BTA-2-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, ~{N},~{N}-dimethyl-4-(6-methyl-1,3-benzothiazol-2-yl)aniline | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5W9S
 
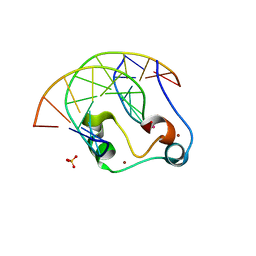 | | Zinc finger of human CXXC5 in complex with CpG DNA | | Descriptor: | CXXC-type zinc finger protein 5, CpG DNA fragment, SULFATE ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
7XAD
 
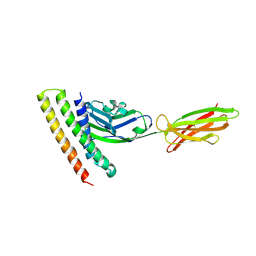 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XAE
 
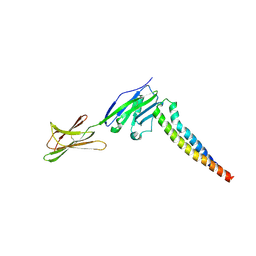 | | Crystal strucutre of PD-L1 and 3ONJA protein | | Descriptor: | 2IC6, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal strucutre of PD-L1 and 2IC6 protein
To Be Published
|
|
6PI7
 
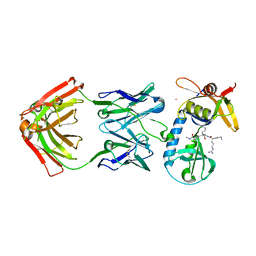 | |
5GSX
 
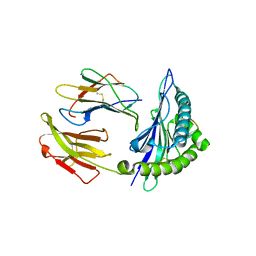 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSR
 
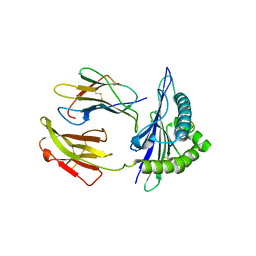 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSV
 
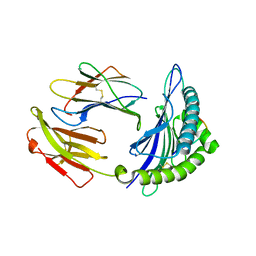 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
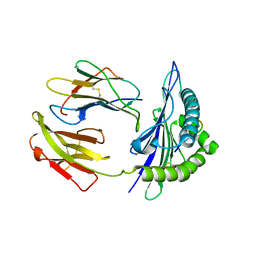 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSB
 
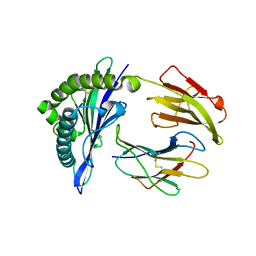 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
7MWK
 
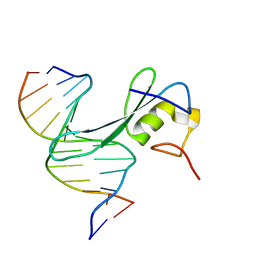 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(MC)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
7MWM
 
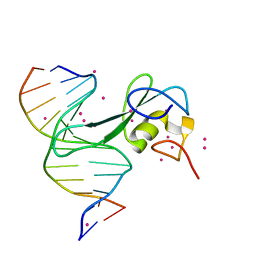 | | Crystal structure of MBD2 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*A)-R(P*(5MC))-D(P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of MBD2 with DNA
To Be Published
|
|
7MWI
 
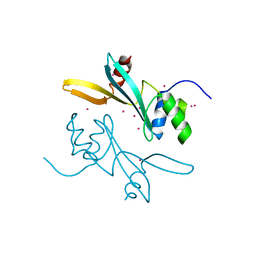 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
7MWL
 
 | | The TAM domain of BAZ2A in complex with a 12mer mCG DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), GLYCEROL | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The TAM domain of BAZ2A in complex with a 12mer mCG DNA
To Be Published
|
|
8X7P
 
 | | CCA-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, copper;trisodium;18-(2-carboxylatoethyl)-20-(carboxylatomethyl)-12-ethenyl-7-ethyl-3,8,13,17-tetramethyl-17,18-dihydroporphyrin-21,23-diide-2-carboxylate | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7R
 
 | | C05-03-bound E46K alpha-synuclein fibrils | | Descriptor: | 2-[(~{E})-4-[6-(methylamino)pyridin-3-yl]but-1-en-3-ynyl]-1,3-benzothiazol-6-ol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7Q
 
 | | pFTAA-bound E46K alpha-synuclein fibrils | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7O
 
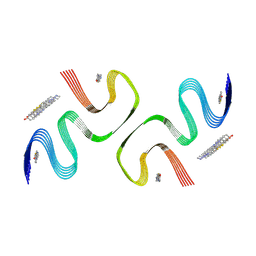 | | PiB-bound E46K mutanted alpha-synuclein fibrils | | Descriptor: | 2-[4-(methylamino)phenyl]-1,3-benzothiazol-6-ol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7M
 
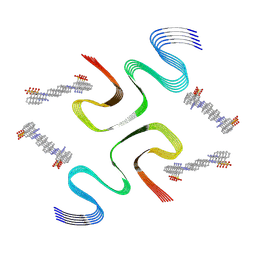 | | CR-bound E46K alpha-synuclein fibrils | | Descriptor: | 4-azanyl-3-[(~{E})-[4-[4-[(~{E})-(1-azanyl-4-sulfo-naphthalen-2-yl)diazenyl]phenyl]phenyl]diazenyl]naphthalene-1-sulfonic acid, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2023-11-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
