2WRD
 
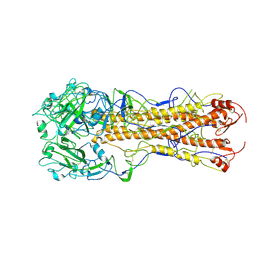 | | structure of H2 japan hemagglutinin | | Descriptor: | HEMAGGLUTININ | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WRH
 
 | | structure of H1 duck albert hemagglutinin with human receptor | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN, N-acetyl-alpha-neuraminic acid | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WR3
 
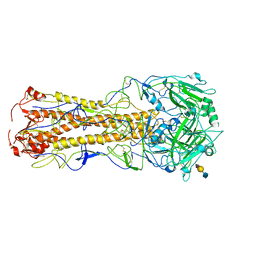 | | structure of influenza H2 duck Ontario hemagglutinin with avian receptor | | Descriptor: | HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WRG
 
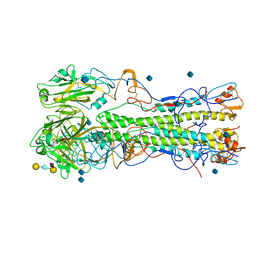 | | structure of H1 1918 hemagglutinin with human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN, ... | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3C2G
 
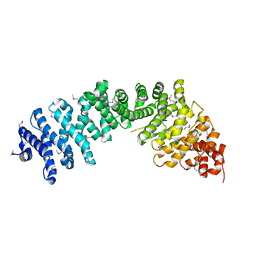 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
2F17
 
 | | Mouse Thiamin Pyrophosphokinase in a Ternary Complex with Pyrithiamin Pyrophosphate and AMP at 2.5 angstrom | | Descriptor: | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-METHYLPYRIDINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Liu, J.Y, Timm, D.E, Hurley, T.D. | | Deposit date: | 2005-11-14 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrithiamine as a substrate for thiamine pyrophosphokinase
J.Biol.Chem., 281, 2006
|
|
3C2H
 
 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
5HXL
 
 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
4X3Q
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SibL | | Authors: | liu, J.S, Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH
To Be Published, 2015
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J3U
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
8GK3
 
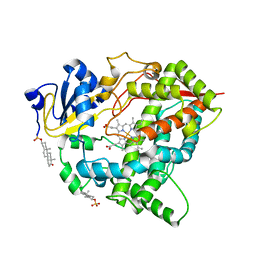 | | Cytochrome P450 3A7 in complex with Dehydroepiandrosterone sulfate | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, Cytochrome P450 3A7, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human cytochrome P450 3A7 binding four copies of its native substrate dehydroepiandrosterone 3-sulfate.
J.Biol.Chem., 299, 2023
|
|
8E7X
 
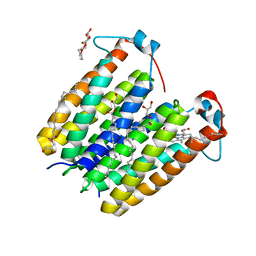 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Z
 
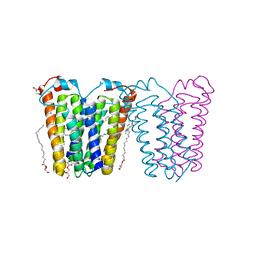 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
3I6G
 
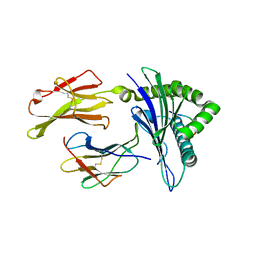 | | Newly identified epitope Mn2 from SARS-CoV M protein complexed withHLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes.
J.INFECT.DIS., 202, 2010
|
|
3H01
 
 | |
2DFS
 
 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
3H00
 
 | |
8F5Q
 
 | | Crystal structure of human PCNA in complex with the PIP box of FBH1 | | Descriptor: | F-box DNA helicase 1, Proliferating cell nuclear antigen | | Authors: | Liu, J, Chaves-Arquero, B, Wei, P, Tencer, H, Zhang, G, Blanco, F, Kutateladze, T. | | Deposit date: | 2022-11-15 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insight into the PCNA-binding mode of FBH1.
Structure, 31, 2023
|
|
1SJJ
 
 | |
5YJ8
 
 | |
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
