3ZP0
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZPA
 
 | | INFLUENZA VIRUS (VN1194) H5 I155F mutant HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ | | Authors: | Liu, J, Chen, Z, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
5OXF
 
 | |
5OWV
 
 | |
3C2G
 
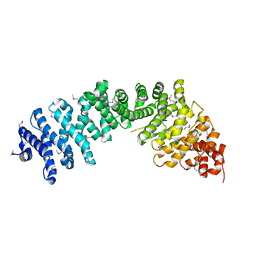 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3C2H
 
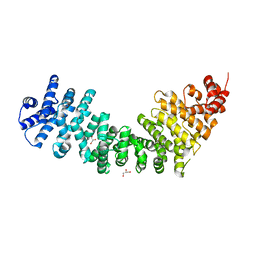 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
7LKY
 
 | |
7LYX
 
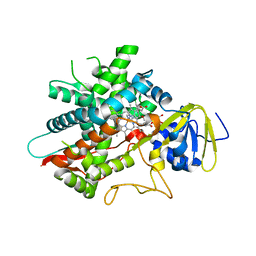 | | Crystal structure of human CYP8B1 in complex with (S)-tioconazole | | Descriptor: | (S)-Tioconazole, 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase, GLYCEROL, ... | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and characterization of human cytochrome P450 8B1 supports future drug design for nonalcoholic fatty liver disease and diabetes.
J.Biol.Chem., 298, 2022
|
|
6U2S
 
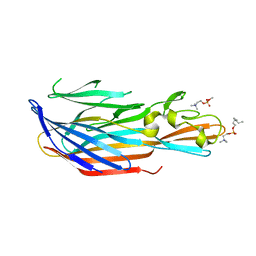 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U33
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, NICKEL (II) ION | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3F
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Panton-Valentine Leucocidin F, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3T
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3Y
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | ACETATE ION, Gamma-hemolysin subunit A, Panton-Valentine Leucocidin F, ... | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WVA
 
 | |
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
5WUL
 
 | |
8K4H
 
 | | Crystal structure of PDE4D complexed with benzbromarone | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K4C
 
 | | Crystal structure of PDE4D complexed with ethaverine hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3,4-diethoxyphenyl)methyl]-6,7-diethoxy-isoquinoline, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
6V9C
 
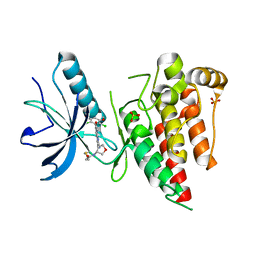 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | Authors: | Liu, J, Liu, H. | | Deposit date: | 2019-12-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
9BA4
 
 | | Full-length cross-linked Contactin 2 (CNTN2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of contactin 2 homophilic interaction.
Structure, 32, 2024
|
|
9BA5
 
 | | Cross-linked Contactin 2 Ig1-Ig6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2 | | Authors: | Liu, J.L, Fan, S.F, Ren, G.R, Rudenko, G.R. | | Deposit date: | 2024-04-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Molecular mechanism of contactin 2 homophilic interaction.
Structure, 32, 2024
|
|
6O1O
 
 | |
