8XYW
 
 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYS
 
 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYT
 
 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVT
 
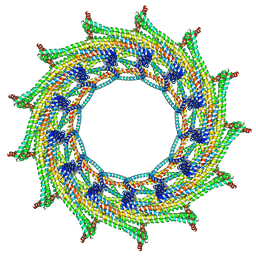 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
7AX3
 
 | |
4YJ5
 
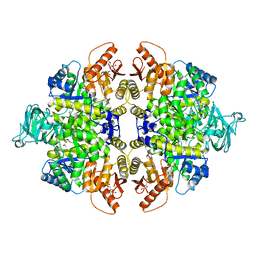 | | Crystal structure of PKM2 mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
7NHR
 
 | |
6WXJ
 
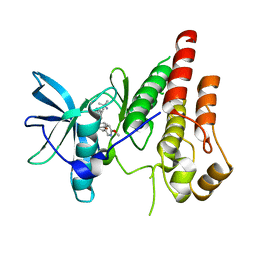 | | CSF1R signaling is a regulator of pathogenesis in progressive MS | | Descriptor: | 4-(3-{[(2S)-2-(6-methoxypyridin-3-yl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl}-3H-imidazo[4,5-b]pyridin-6-yl)-2-methylbut-3-yn-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | CSF1R signaling is a regulator of pathogenesis in progressive MS.
Cell Death Dis, 11, 2020
|
|
6WW3
 
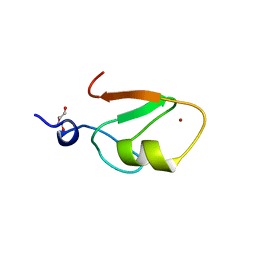 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
4QVG
 
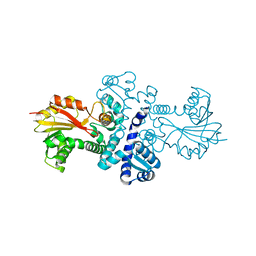 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in its apo form | | Descriptor: | SibL | | Authors: | Liu, J.S, Chen, S.C, Huang, C.H, Yang, C.S, Chen, Y. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of an antibiotics-synthesizing 3-hydroxykynurenine C-methyltransferase
Sci Rep, 5, 2015
|
|
1P2A
 
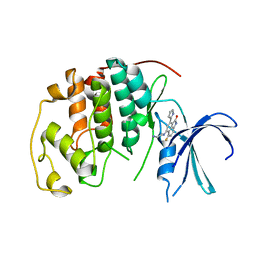 | | The structure of cyclin dependent kinase 2 (CKD2) with a trisubstituted naphthostyril inhibitor | | Descriptor: | 5-[(2-AMINOETHYL)AMINO]-6-FLUORO-3-(1H-PYRROL-2-YL)BENZO[CD]INDOL-2(1H)-ONE, Cell division protein kinase 2 | | Authors: | Liu, J.-J, Dermatakis, A, Lukacs, C.M, Konzelmann, F, Chen, Y, Kammlott, U, Depinto, W, Yang, H, Yin, X, Chen, Y, Schutt, A, Simcox, M.E, Luk, K.-C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3,5,6-Trisubstituted Naphthostyrils as CDK2 Inhibitors
BIOORG.MED.CHEM., 13, 2003
|
|
6PW1
 
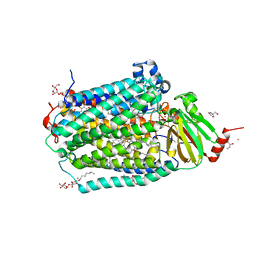 | | Cytochrome c Oxidase delta 16 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
6PW0
 
 | | Cytochrome C oxidase delta 6 mutant | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
4MJ5
 
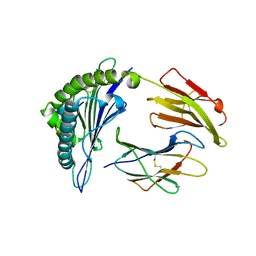 | | Crystal Structure of HLA-A*1101 in complex with H1-22, an influenza A(H1N1) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
4MJ6
 
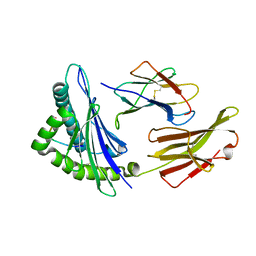 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
1I5Y
 
 | | HIV-1 GP41 CORE | | Descriptor: | SULFATE ION, TRANSMEMBRANE GLYCOPROTEIN (GP41) | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-03-01 | | Release date: | 2002-09-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of interhelical interactions in the human
immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning
mutagenesis.
J.Virol., 75, 2001
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
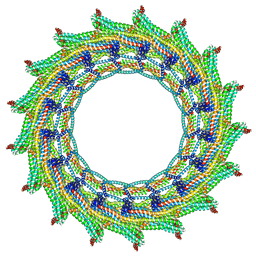 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
4X3Q
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SibL | | Authors: | liu, J.S, Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH
To Be Published, 2015
|
|
