5YI2
 
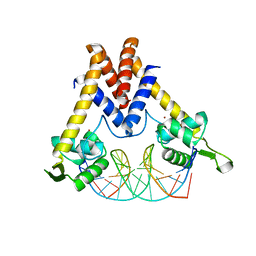 | | Structure of Lactococcus lactis ZitR, wild type in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
4EAY
 
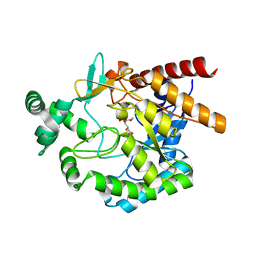 | | Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate | | Descriptor: | CHLORIDE ION, D-MANNONIC ACID, MANGANESE (II) ION, ... | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
4EAC
 
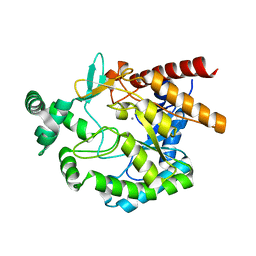 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
5ZTL
 
 | | Non-cryogenic structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, S.Y, Liu, H, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
3SJ8
 
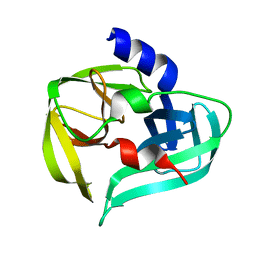 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJI
 
 | | crystal structure of CVA16 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, SODIUM ION | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJ9
 
 | | crystal structure of the C147A mutant 3C of CVA16 in complex with FAGLRQAVTQ peptide | | Descriptor: | 3C protease, FAGLRQAVTQ peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
7V6H
 
 | | Crystal Structure of the SpnL | | Descriptor: | Cyclopropane fatty-acyl-phospholipid synthase-like methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, H.-H, Ko, T.-P, Liu, H.-W, Tsai, M.-D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Evidence for an Enzyme-Catalyzed Rauhut-Currier Reaction during the Biosynthesis of Spinosyn A.
J.Am.Chem.Soc., 143, 2021
|
|
4J2T
 
 | | Inhibitor-bound Ca2+ ATPase | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9bS)-6-(acetyloxy)-3a,4-bis(butanoyloxy)-3-hydroxy-3,6,9-trimethyl-8-{[(2E)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-7-yl octanoate, PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, ... | | Authors: | Paulsen, E.S, Villadsen, J, Tenori, E, Liu, H, Lie, M.A, Bonde, D.F, Bublitz, M, Olesen, C, Autzen, H.E, Dach, I, Sehgal, P, Moller, J.V, Schiott, B, Nissen, P, Christensen, S.B. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase.
J.Med.Chem., 56, 2013
|
|
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-18 | | Last modified: | 2018-09-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
3ZIF
 
 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
2IX2
 
 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5YT7
 
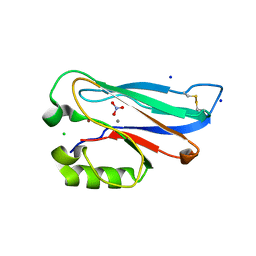 | |
2JG2
 
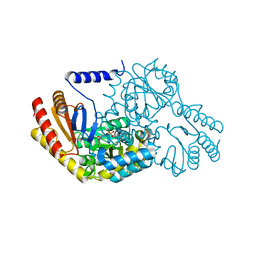 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
2JVN
 
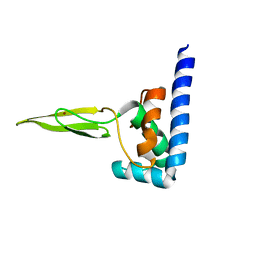 | | Domain C of human PARP-1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Hoffman, D, Tao, Z, Liu, H. | | Deposit date: | 2007-09-24 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif.
Biochemistry, 47, 2008
|
|
2JGT
 
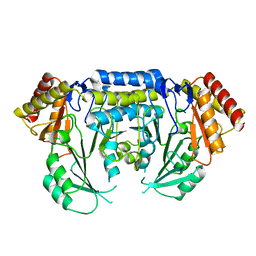 | | Low resolution structure of SPT | | Descriptor: | SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
4MBS
 
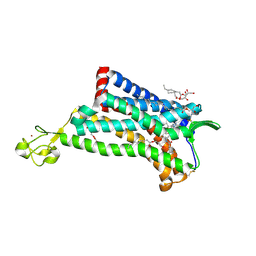 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
5WSP
 
 | | Crystal structure of DNA3 duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), STRONTIUM ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSR
 
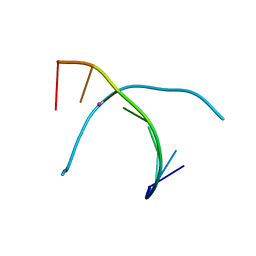 | | Crystal structure of T-Hg-T pair containing DNA duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSQ
 
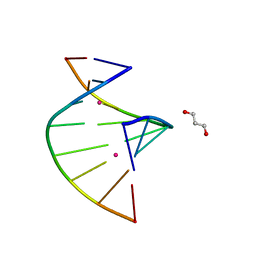 | | Crystal structure of C-Hg-T pair containing DNA duplex | | Descriptor: | 1,3-PROPANDIOL, DNA (5'-D(*GP*CP*CP*CP*GP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSS
 
 | |
4MQW
 
 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor (P31) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Follicle-stimulating hormone receptor, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2013-09-16 | | Release date: | 2014-04-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence for Follicle-stimulating Hormone Receptor as a Functional Trimer.
J.Biol.Chem., 289, 2014
|
|
1F1G
 
 | | Crystal structure of yeast cuznsod exposed to nitric oxide | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, PHOSPHATE ION, ... | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
