6XKQ
 
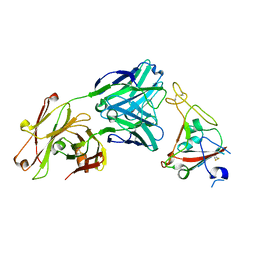 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
1MYQ
 
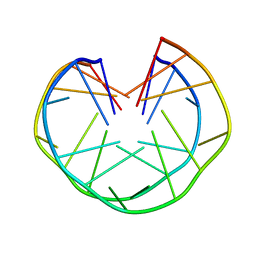 | | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction | | Descriptor: | 5'-D(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Matsugami, A, Ouhashi, K, Kanagawa, M, Liu, H, Kanagawa, S, Uesugi, S, Katahira, M. | | Deposit date: | 2002-10-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction.
J.Mol.Biol., 313, 2001
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
5WSP
 
 | | Crystal structure of DNA3 duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), STRONTIUM ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSR
 
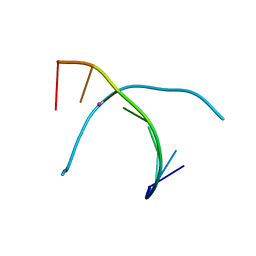 | | Crystal structure of T-Hg-T pair containing DNA duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
3VPN
 
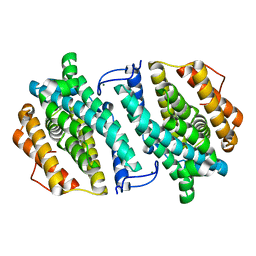 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPM
 
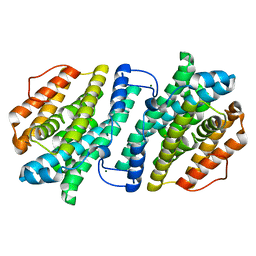 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
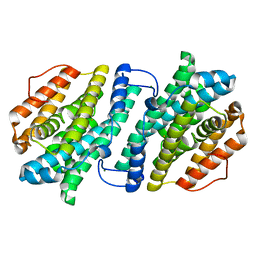 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
4GXB
 
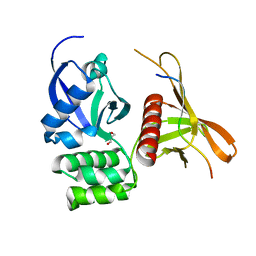 | | Structure of the SNX17 atypical FERM domain bound to the NPxY motif of P-selectin | | Descriptor: | GLYCEROL, P-selectin, Sorting nexin-17 | | Authors: | Ghai, R, Bugarcic, A, Liu, H, Norwood, S.J, Li, S.S, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2012-09-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
1FOR
 
 | |
1F81
 
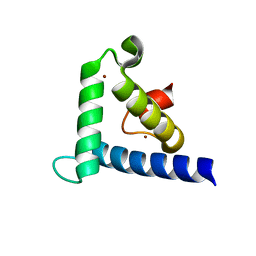 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
7DGY
 
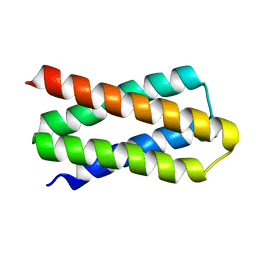 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6LZE
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11a | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhang, Y, Jing, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6E67
 
 | | Structure of beta2 adrenergic receptor fused to a Gs peptide | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor,Endolysin,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Beta-2 adrenergic receptor chimera | | Authors: | Liu, X, Xu, X, Hilger, D, Tiemann, J, Liu, H, Du, Y, Hirata, K, Sun, X, Guixa-Gonzalez, R, Mathiesen, J, Hildebrand, P, Kobilka, B. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
6XWK
 
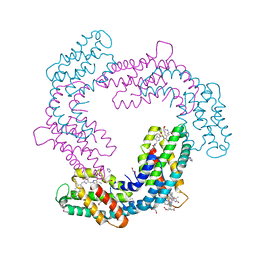 | | Crystal structure of Phormidium rubidum phycocyanin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Sonani, R.R, Roszak, A.W, Cogdell, R.J, Madamwar, D, Liu, H, Gross, M.L, Blankenship, R.E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Photosyn. Res., 144, 2020
|
|
4R99
 
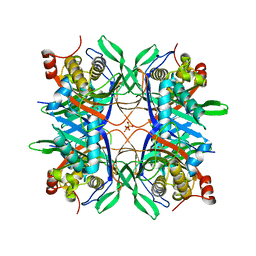 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6L2L
 
 | |
6L2M
 
 | |
6OMM
 
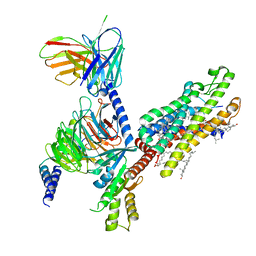 | | Cryo-EM structure of formyl peptide receptor 2/lipoxin A4 receptor in complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Liu, H, de Waal, P.W, Zhou, X.E, Wang, L, Meng, X, Zhao, G, Kang, Y, Melcher, K, Xu, H.E, Zhang, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of formylpeptide receptor 2-Gicomplex reveals insights into ligand recognition and signaling.
Nat Commun, 11, 2020
|
|
1ORR
 
 | |
2X5F
 
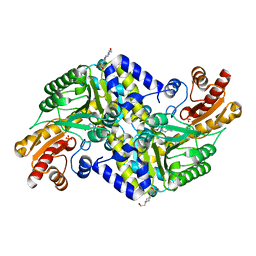 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
