7PEG
 
 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6FI7
 
 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GUM
 
 | | Structure of the A.thaliana E1 UFD domain in complex with E2 | | Descriptor: | GLYCEROL, SAE2, SUMO-conjugating enzyme SCE1 | | Authors: | Liu, B, Lois, L.M, Reverter, D. | | Deposit date: | 2018-06-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into SUMO E1-E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution.
Biochem.J., 476, 2019
|
|
6GV3
 
 | |
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
9CV6
 
 | |
9CGI
 
 | |
8JVA
 
 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | Authors: | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC2
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBY
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBZ
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC4
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC1
 
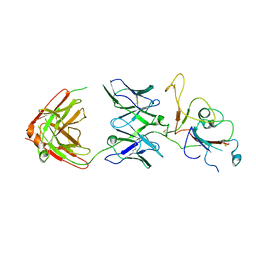 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC6
 
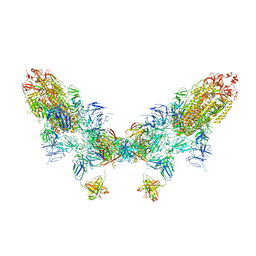 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.85 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
5LGM
 
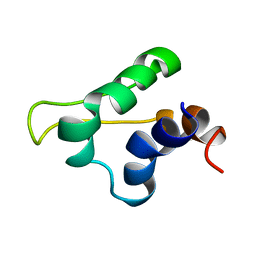 | | Gp5.7 mutant L42A | | Descriptor: | Fusion protein 5.5/5.7 | | Authors: | Liu, B, Matthews, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gp5.7 mutant L42A
to be published
|
|
6PMJ
 
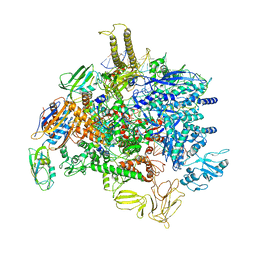 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6PMI
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
1UDE
 
 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
8IVH
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-27 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | crystal structure of SulE mutant
To Be Published
|
|
8J7L
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | crystal structure of SulE mutant
To Be Published
|
|
