2M7P
 
 | | RXFP1 utilises hydrophobic moieties on a signalling surface of the LDLa module to mediate receptor activation | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Relaxin receptor 1 | | Authors: | Kong, R.CK, Petrie, E.J, Mohanty, B, Ling, J, Lee, J.C.Y, Gooley, P.R, Bathgate, R.A.D. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.
J.Biol.Chem., 288, 2013
|
|
1WAK
 
 | | X-ray structure of SRPK1 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE SPRK1 | | Authors: | Ngo, J.C, Gullingsrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.D, Adams, J.A, Mccammon, J.A, Ghosh, G. | | Deposit date: | 2004-10-26 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Sr Protein Kinase 1 is Resilient to Inactivation.
Structure, 15, 2007
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
2GC8
 
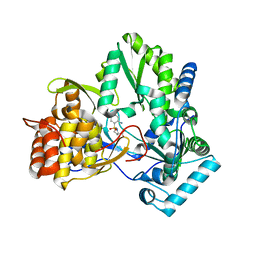 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
4W4T
 
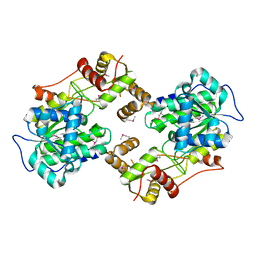 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
4PJS
 
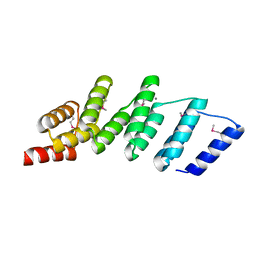 | | Crystal structure of designed (SeMet)-cPPR-NRE protein | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJQ
 
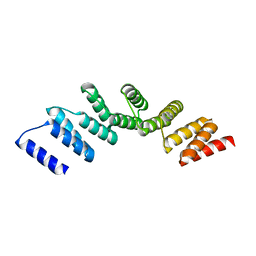 | | Crystal structure of designed cPPR-polyG protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJR
 
 | | Crystal structure of designed cPPR-NRE protein | | Descriptor: | MAGNESIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
1YBJ
 
 | | Structural and Dynamics studies of both apo and holo forms of the hemophore HasA | | Descriptor: | Hemophore HasA | | Authors: | Wolff, N, Izadi-Pruneyre, N, Couprie, J, Habeck, M, Linge, J, Rieping, W, Wandersman, C, Nilges, M, Delepierre, M, Lecroisey, A. | | Deposit date: | 2004-12-21 | | Release date: | 2005-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications.
J.Mol.Biol., 376, 2008
|
|
4GKR
 
 | |
4GKQ
 
 | |
4GUB
 
 | | Casein Kinase II bound to Inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, N-[5-({3-cyano-7-[(1-methyl-1H-imidazol-4-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl}amino)-2-methylphenyl]acetamide | | Authors: | Larsen, N.A, Dowling, J.E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and selective inhibitors of CK2 kinase.
To be Published
|
|
7AW0
 
 | | MerTK kinase domain in complex with purine inhibitor | | Descriptor: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
1D8B
 
 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
1KKS
 
 | | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression | | Descriptor: | 5'-R(*GP*GP*AP*AP*GP*GP*CP*CP*CP*UP*UP*UP*UP*CP*AP*GP*GP*GP*CP*CP*AP*CP*CP*C)-3' | | Authors: | Zanier, K, Luyten, I, Crombie, C, Muller, B, Schuemperli, D, Linge, J.P, Nilges, M, Sattler, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression.
RNA, 8, 2002
|
|
6PRY
 
 | | X-ray crystal structure of the blue-light absorbing state of PixJ from Thermosynechococcus elongatus by serial femtosecond crystallographic analysis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M, Kern, J.F. | | Deposit date: | 2019-07-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PRU
 
 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2019-07-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FBC
 
 | | Crystal structure of P450T2 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pereira, J.H, Huang, J, Keasling, J, Adams, P.D. | | Deposit date: | 2022-11-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Complete integration of carbene-transfer chemistry into biosynthesis.
Nature, 617, 2023
|
|
6OBV
 
 | | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases | | Descriptor: | fluvirucin B1 DH domain from module 1 | | Authors: | McAndrew, R.P, Barajas, J.F, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases.
J Ind Microbiol Biotechnol., 46, 2019
|
|
6OBT
 
 | | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases | | Descriptor: | Borrelidin polyketide synthase, type I | | Authors: | McAndrew, R.P, Barajas, J.F, Pereira, J.H, Keasling, J.D, Adams, P.D. | | Deposit date: | 2019-03-21 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases.
J Ind Microbiol Biotechnol., 46, 2019
|
|
8A00
 
 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
5MG1
 
 | | Structure of the photosensory module of Deinococcus phytochrome by serial femtosecond X-ray crystallography | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Young, I.D, Brewster, A.S, Clinger, J, Andi, B, Stan, C, Allaire, M, Nelsen, S, Alonso-Mori, R, Phillips Jr, G.N, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
