1TJZ
 
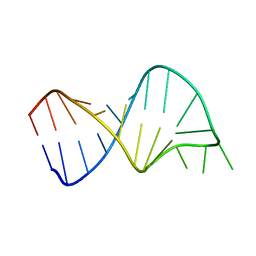 | |
1Z2T
 
 | | NMR structure study of anchor peptide Ser65-Leu87 of enzyme acholeplasma laidlawii Monoglycosyldiacyl Glycerol Synthase (alMGS) in DHPC micelles | | Descriptor: | Anchor peptide Ser65-Leu87 of alMGS | | Authors: | Lind, J, Barany-Wallje, E, Ramo, T, Wieslander, A, Maler, L. | | Deposit date: | 2005-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, position of and membrane-interaction of a putative membrane-anchoring domain of alMGS
To be Published
|
|
6NUJ
 
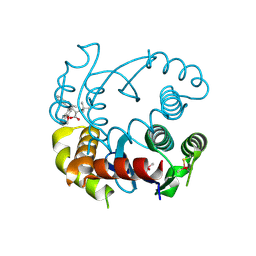 | |
6EB1
 
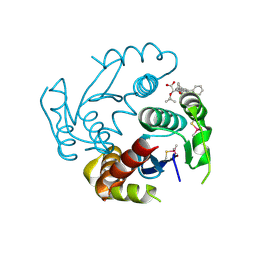 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6EB2
 
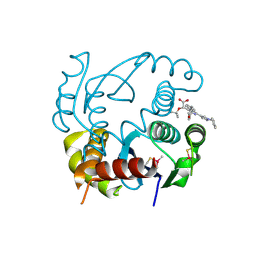 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid | | Descriptor: | (2S)-[1-(1-benzyl-1H-pyrazol-4-yl)-3-(3,4-dihydro-2H-1-benzopyran-6-yl)isoquinolin-4-yl](tert-butoxy)acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
8FX9
 
 | |
6PU1
 
 | |
8GDO
 
 | |
7KE0
 
 | |
1HWQ
 
 | |
1Q8N
 
 | | Solution Structure of the Malachite Green RNA Binding Aptamer | | Descriptor: | MALACHITE GREEN, RNA Aptamer | | Authors: | Flinders, J, DeFina, S.C, Brackett, D.M, Baugh, C, Wilson, C, Dieckmann, T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex.
Chembiochem, 5, 2004
|
|
1JQS
 
 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQT
 
 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | Descriptor: | 50S Ribosomal protein L11 | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1MVR
 
 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | Descriptor: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | Authors: | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1MI6
 
 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1JQM
 
 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
2L7C
 
 | | Biophysical studies of lipid interacting regions of DGD2 in Arabidopsis thaliana | | Descriptor: | Digalactosyldiacylglycerol synthase 2, chloroplastic | | Authors: | Szpryngiel, S, Ge, C, Iakovleva, I, Lind, J, Wieslander, A, Maler, L. | | Deposit date: | 2010-12-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lipid interacting regions in phosphate stress glycosyltransferase atDGD2 from Arabidopsis thaliana.
Biochemistry, 50, 2011
|
|
2K44
 
 | |
4UMQ
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-{5-[(3-hydroxy-5-methoxyphenyl)amino]-2-(phenylcarbamoyl)phenoxy}propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMT
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMR
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4U31
 
 | | Sco GlgEI-V279S in Complex with maltose-C-phosphonate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U3C
 
 | | Docking Site of Maltohexaose in the Mtb GlgE | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
