6LZ6
 
 | |
6LZ8
 
 | |
4Q7Z
 
 | | Neutrophil serine protease 4 (PRSS57) with phe-phe-arg-chloromethylketone (FFR-cmk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4Q80
 
 | | Neutrophil serine protease 4 (PRSS57) with val-leu-lys-chloromethylketone (VLK-cmk) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-valyl-N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-L-leucinamide, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4Q7Y
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
4Q7X
 
 | | Neutrophil serine protease 4 (PRSS57) apo form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
8GYB
 
 | |
8GY9
 
 | |
8ASA
 
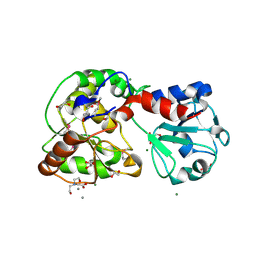 | | Crystal structure of AO75L | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of AO75L
To Be Published
|
|
8AVQ
 
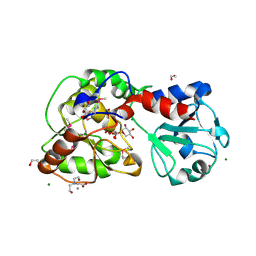 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AO75L in Complex with UDP-Xylose
To Be Published
|
|
1MVP
 
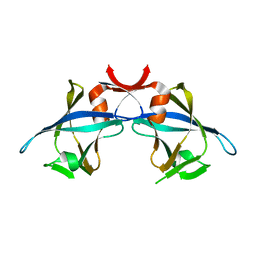 | |
6MNE
 
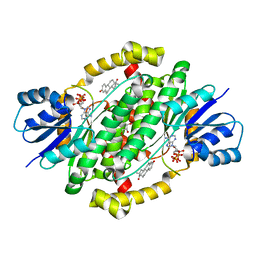 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE AND NADP+ | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estradiol 17-beta-dehydrogenase 1, GLYCEROL, ... | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
6MNC
 
 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, DI(HYDROXYETHYL)ETHER, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Li, T, Stephen, P, Zhu, D.W, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
5AJW
 
 | | Human PFKFB3 in complex with an indole inhibitor 2 | | Descriptor: | 2-amino-N-[4-(2-amino-1-benzyl-3-cyano-indol-5-yl)oxyphenyl]acetamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5AJZ
 
 | | Human PFKFB3 in complex with an indole inhibitor 5 | | Descriptor: | 2-azanyl-N-[4-[(3-cyano-1H-indol-5-yl)oxy]phenyl]ethanamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5AK0
 
 | | Human PFKFB3 in complex with an indole inhibitor 6 | | Descriptor: | (2S)-N-[4-[1-METHYL-3-(1-METHYLPYRAZOL-4-YL)INDOL-5-YL]OXYPHENYL]PYRROLIDINE-2-CARBOXAMIDE, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5B63
 
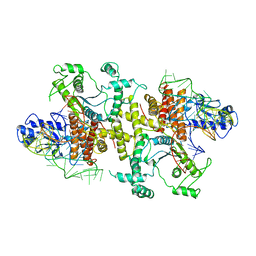 | | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, tRNA-Arg | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg)
To Be Published
|
|
3CKZ
 
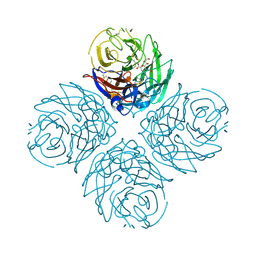 | | N1 Neuraminidase H274Y + Zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR | | Authors: | Colllins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3CL2
 
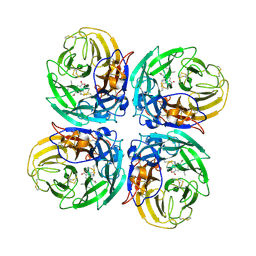 | | N1 Neuraminidase N294S + Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
5A4F
 
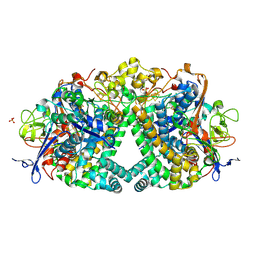 | | The mechanism of Hydrogen Activation by NiFe-hydrogenases. | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
2YP7
 
 | | Haemagglutinin of 2005 Human H3N2 Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Xiong, X, Lin, Y.P, Wharton, S.A, Martin, S.R, Coombs, P.J, Vachieri, S.G, Christodoulou, E, Walker, P.A, Liu, J, Skehel, J.J, Gamblin, S.J, Hay, A.J, Daniels, R.S, McCauley, J.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of the Receptor Binding Properties of the Influenza A(H3N2) Hemagglutinin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5A7D
 
 | | Tetrameric assembly of LGN with Inscuteable | | Descriptor: | INSCUTEABLE, PINS | | Authors: | Culurgioni, S, Mari, S, Bonetto, G, Gallini, S, Brennich, M, Round, A, Mapelli, M. | | Deposit date: | 2015-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of the Insc:Lgn Tetramer Reveals a New Function of Lgn in Promoting Asymmetric Cell Divisions
To be Published
|
|
