9EV3
 
 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO) purified from bacteria grown in acetate minimal medium | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
7C4H
 
 | |
9EV4
 
 | | Pyruvate:quinone oxidoreductase (PQO) from Corynebacterium glutamicum CS176 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thiamine pyrophosphate-requiring enzymes [acetolactate synthase, pyruvate dehydrogenase (Cytochrome), ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
9EV6
 
 | | Corynebacterium glutamicum pyruvate:quinone oxidoreductase (PQO), C-terminal truncated construct | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Da Silva Lameira, C, Muenssinger, S, Yang, L, Eikmanns, B.J, Bellinzoni, M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Corynebacterium glutamicum pyruvate:quinone oxidoreductase: an enigmatic metabolic enzyme with unusual structural features.
Febs J., 291, 2024
|
|
3BIV
 
 | | Human thrombin-in complex with UB-THR11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
7DUT
 
 | | Structure of Sulfolobus solfataricus SegA protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DUV
 
 | | Structure of Sulfolobus solfataricus SegB protein | | Descriptor: | SULFATE ION, SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV2
 
 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV3
 
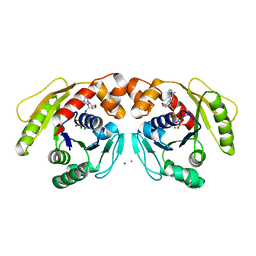 | | Structure of Sulfolobus solfataricus SegA-AMPPNP protein | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Wu, C.T, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
4I5M
 
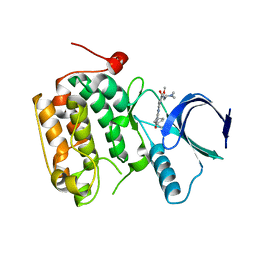 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
6EFK
 
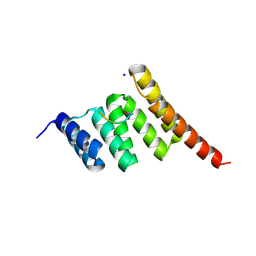 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
4MO2
 
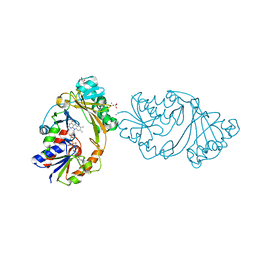 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
7RDB
 
 | | Crystal structure of Tspan15 large extracellular loop (Tspan15 LEL) | | Descriptor: | Tetraspanin-15 | | Authors: | Lipper, C.H, Gabriel, K.H, Seegar, T.C.M, Durr, K.L, Tomlinson, M.G, Blacklow, S.C. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the Tspan15 LEL domain reveals a conserved ADAM10 binding site.
Structure, 30, 2022
|
|
6EDI
 
 | |
7QDX
 
 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
7RD5
 
 | | Crystal structure of Tspan15 large extracellular loop (Tspan15 LEL) in complex with 1C12 Fab | | Descriptor: | 1C12 Fab Heavy Chain, 1C12 Fab Light Chain, Tetraspanin-15 | | Authors: | Lipper, C.H, Gabriel, K.H, Seegar, T.C.M, Durr, K.L, Tomlinson, M.G, Blacklow, S.C. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the Tspan15 LEL domain reveals a conserved ADAM10 binding site.
Structure, 30, 2022
|
|
7Q9W
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 4-(aminomethyl)pyridin-2-amine | | Descriptor: | 4-(aminomethyl)pyridin-2-amine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QA7
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Cyclopropanemethylamine | | Descriptor: | Cholinephosphate cytidylyltransferase, cyclopropylmethanamine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7Q9V
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1-(1,3-oxazol-4-yl)methanamine | | Descriptor: | 1,3-oxazol-4-ylmethanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QAD
 
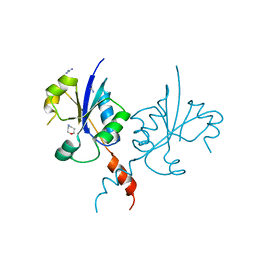 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1,4-Oxazepane hydrochloride | | Descriptor: | 1,4-oxazepane, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
7QD3
 
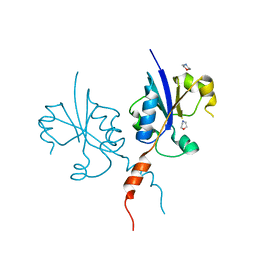 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with morpholine | | Descriptor: | Cholinephosphate cytidylyltransferase, morpholine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QVO
 
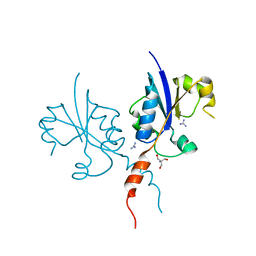 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with guanidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QVN
 
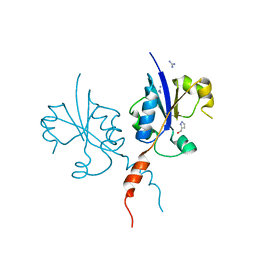 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with(1H-pyrazol-5-yl)methanol | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7QBJ
 
 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
