4PMX
 
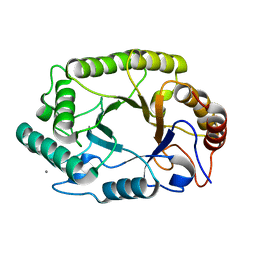 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form | | Descriptor: | CALCIUM ION, Xylanase | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
2B4U
 
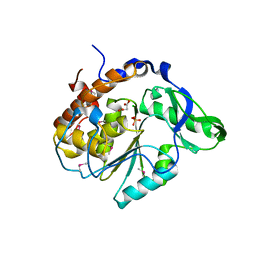 | | Structure of the C252S mutant of Selenomonas ruminantium PTP-like phytase | | Descriptor: | CHLORIDE ION, MALONATE ION, SULFATE ION, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2005-09-26 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of a bacterial protein tyrosine phosphatase-like myo-inositol polyphosphatase.
Protein Sci., 16, 2007
|
|
2B4P
 
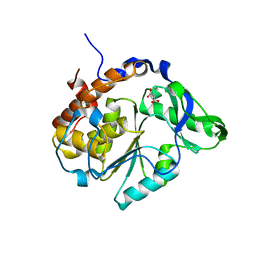 | |
4R7Z
 
 | | PfMCM-AAA double-octamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 21, MAGNESIUM ION | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
3U2G
 
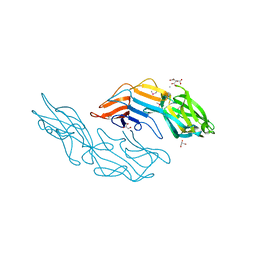 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2QMT
 
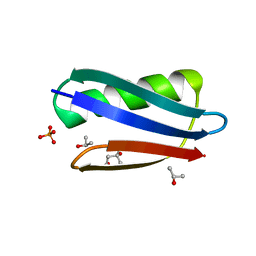 | | Crystal Polymorphism of Protein GB1 Examined by Solid-state NMR and X-ray Diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOPROPYL ALCOHOL, Immunoglobulin G-binding protein G, ... | | Authors: | Frericks Schmidt, H.L, Sperling, L.J, Gao, Y.G, Wylie, B.J, Boettcher, J.M, Wilson, S.R, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Polymorphism of Protein GB1 Examined by Solid-State NMR Spectroscopy and X-ray Diffraction.
J.Phys.Chem.B, 111, 2007
|
|
1ENN
 
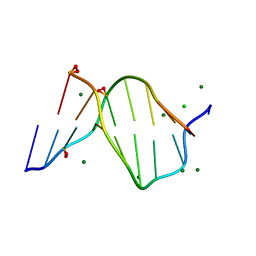 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
1EXD
 
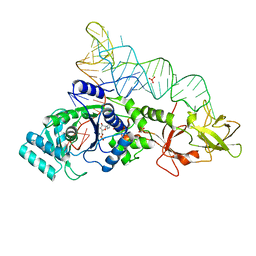 | | CRYSTAL STRUCTURE OF A TIGHT-BINDING GLUTAMINE TRNA BOUND TO GLUTAMINE AMINOACYL TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMINE TRNA APTAMER, GLUTAMINYL-TRNA SYNTHETASE, ... | | Authors: | Bullock, T.L, Sherlin, L.D, Perona, J.J. | | Deposit date: | 2000-05-02 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary core rearrangements in a tight binding transfer RNA aptamer.
Nat.Struct.Biol., 7, 2000
|
|
2LTQ
 
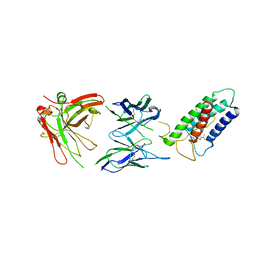 | | High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Tang, M, Sperling, L.J, Schwieters, C.D, Nesbitt, A.E, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer.
J.Mol.Biol., 425, 2013
|
|
2QW1
 
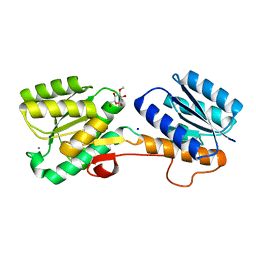 | | Glucose/galactose binding protein bound to 3-O-methyl D-glucose | | Descriptor: | 3-O-methyl-beta-D-glucopyranose, CALCIUM ION, D-galactose-binding periplasmic protein, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2007-08-09 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a periplasmic binding protein antagonist that prevents domain closure.
Acs Chem.Biol., 4, 2009
|
|
1ETJ
 
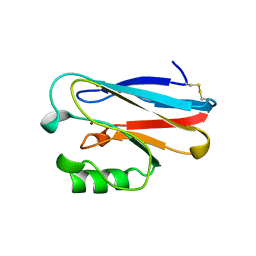 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLU | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Karlsson, B.G, Tsai, L.-C, Nar, H, Sanders-Loehr, J, Bonander, N, Langer, V, Sjolin, L. | | Deposit date: | 1997-01-11 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure determination and characterization of the Pseudomonas aeruginosa azurin mutant Met121Glu.
Biochemistry, 36, 1997
|
|
4BZU
 
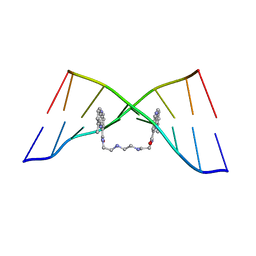 | | The Solution Structure of the MLN 944-d(TATGCATA)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
1EZL
 
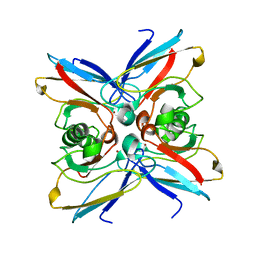 | | CRYSTAL STRUCTURE OF THE DISULPHIDE BOND-DEFICIENT AZURIN MUTANT C3A/C26A: HOW IMPORTANT IS THE S-S BOND FOR FOLDING AND STABILITY? | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Bonander, N, Leckner, J, Guo, H, Karlsson, B.G, Sjolin, L. | | Deposit date: | 2000-05-11 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the disulfide bond-deficient azurin mutant C3A/C26A: how important is the S-S bond for folding and stability?
Eur.J.Biochem., 267, 2000
|
|
2MAI
 
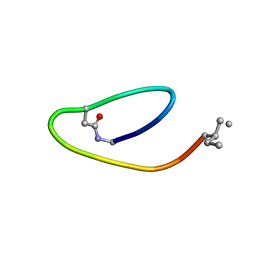 | | NMR structure of lassomycin | | Descriptor: | Lassomycin | | Authors: | Gavrish, E, Sit, C.S, Kandror, O, Spoering, A, Peoples, A, Ling, L, Fetterman, A, Hughes, D, Cao, S, Bissell, A, Torrey, H, Akopian, T, Mueller, A, Epstein, S, Goldberg, A, Clardy, J, Lewis, K. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2.
Chem.Biol., 21, 2014
|
|
4GHS
 
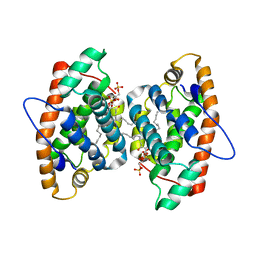 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHP
 
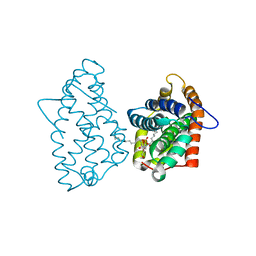 | | Crystal Structure of D48V||A47D mutant of Human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4BZT
 
 | | The Solution Structure of the MLN 944-d(ATGCAT)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
2TSA
 
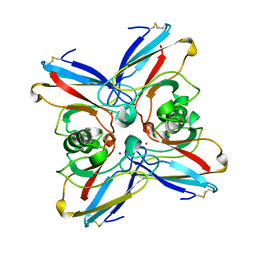 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2M0R
 
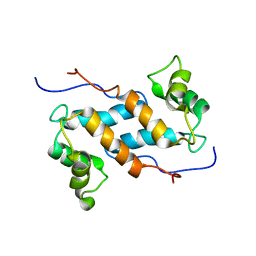 | | Solution structure and dynamics of human S100A14 | | Descriptor: | Protein S100-A14 | | Authors: | Bertini, I, Borsi, V, Cerofolini, L, Das Gupta, S, Fragai, M, Luchinat, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human S100A14.
J.Biol.Inorg.Chem., 18, 2013
|
|
4DHP
 
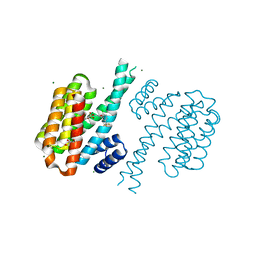 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
473D
 
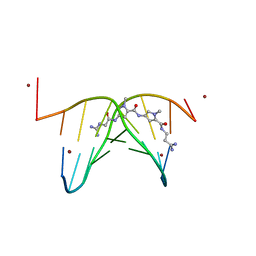 | |
426D
 
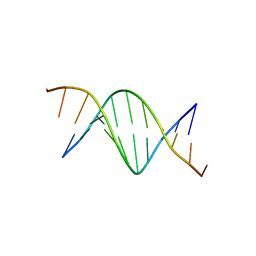 | |
1NZR
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
