7CF9
 
 | | Structure of RyR1 (Ca2+/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
2O8P
 
 | | Crystal structure of a putative 14-3-3 protein from Cryptosporidium parvum, cgd7_2470 | | Descriptor: | 14-3-3 domain containing protein | | Authors: | Dong, A, Lew, J, Wasney, G, Lin, L, Hassanali, A, Zhao, Y, Vedadi, M, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Walker, J.R, Bochkarev, A, Hui, R, Brokx, S.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
7CSP
 
 | | Structure of Ephexin4 IDPSH | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSR
 
 | | Structure of Ephexin4 R676L | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CSO
 
 | | Structure of Ephexin4 DH-PH-SH3 | | Descriptor: | Rho guanine nucleotide exchange factor 16, SULFATE ION | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DE7
 
 | | Crystal structure of PDZD7 HHD domain | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, PDZ domain-containing protein 7 | | Authors: | Wang, H, Lin, L, Lu, Q. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and Membrane Targeting of the PDZD7 Harmonin Homology Domain (HHD) Associated With Hearing Loss.
Front Cell Dev Biol, 9, 2021
|
|
2NPM
 
 | | crystal structure of Cryptosporidium parvum 14-3-3 protein in complex with peptide | | Descriptor: | 14-3-3 domain containing protein, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Dong, A, Lew, J, Wasney, G, Ren, H, Lin, L, Hassanali, A, Qiu, W, Zhao, Y, Doyle, D, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
6J6O
 
 | | Crystal structure of diamondback moth ryanodine receptor phosphorylation domain(2836-3050) | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor, ... | | Authors: | Xu, T, Lin, L, Yuchi, Z. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structure of diamondback moth ryanodine receptor Repeat34 domain reveals insect-specific phosphorylation sites.
Bmc Biol., 17, 2019
|
|
2PBF
 
 | | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative protein-L-isoaspartate O-methyltransferase beta-aspartate methyltransferase (PCMT) from Plasmodium falciparum in complex with S-adenosyl-L-homocysteine
To be Published
|
|
5WKA
 
 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
2N3D
 
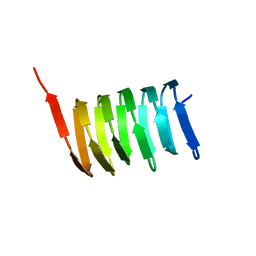 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
2OQK
 
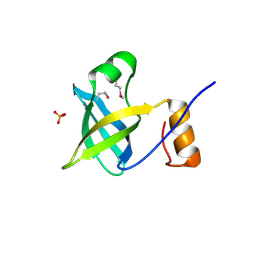 | | Crystal structure of putative Cryptosporidium parvum translation initiation factor eIF-1A | | Descriptor: | Putative translation initiation factor eIF-1A, SULFATE ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Qiu, W, Brokx, S.J, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative Cryptosporidium parvum translation initiation factor eIF-1A
To be Published
|
|
2OWA
 
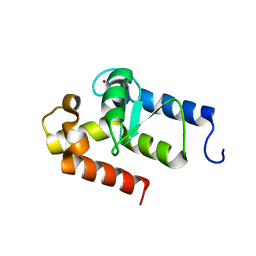 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
2ONU
 
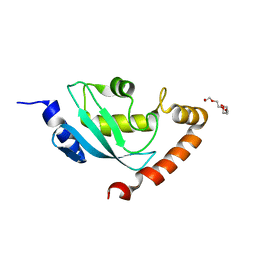 | | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H | | Descriptor: | TETRAETHYLENE GLYCOL, Ubiquitin-conjugating enzyme, putative | | Authors: | Dong, A, Lew, J, Lin, L, Hassanali, A, Zhao, Y, Senisterra, G, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Hui, R, Brokx, S.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H
To be Published
|
|
2PWQ
 
 | | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii | | Descriptor: | Ubiquitin conjugating enzyme | | Authors: | Qiu, W, Dong, A, Hassanali, A, Lin, L, Brokx, S, Altamentova, S, Hills, T, Lew, J, Ravichandran, M, Kozieradzki, I, Zhao, Y, Schapira, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii.
To be Published
|
|
2POE
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | Descriptor: | Cyclophilin-like protein, putative, FORMIC ACID | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
2PLW
 
 | | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052). | | Descriptor: | Ribosomal RNA methyltransferase, putative, S-ADENOSYLMETHIONINE, ... | | Authors: | Wernimont, A.K, Hassanali, A, Lin, L, Lew, J, Zhao, Y, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a ribosomal RNA methyltransferase, putative, from Plasmodium falciparum (PF13_0052).
To be Published
|
|
4MDP
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens in complex with glucose | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6UNV
 
 | |
8E1X
 
 | | FGFR2 kinase domain in complex with a Pyrazolo[1,5-a]pyrimidine analog (Compound 29) | | Descriptor: | (5M)-N-methyl-5-{(6M,8S)-5-{[(3S)-oxolan-3-yl]amino}-6-[1-(propan-2-yl)-1H-pyrazol-3-yl]pyrazolo[1,5-a]pyrimidin-3-yl}pyridine-3-carboxamide, Fibroblast growth factor receptor 2 | | Authors: | Lei, H.-T, Epling, L.B, Deller, M.C. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Wild-Type and Gatekeeper Mutant Fibroblast Growth Factor Receptor (FGFR) 2/3.
J.Med.Chem., 65, 2022
|
|
1F2H
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE TNFR1 ASSOCIATED PROTEIN, TRADD. | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN | | Authors: | Tsao, D, McDonaugh, T, Malakian, K, Xu, G.-Y, Telliez, J.-B, Hsu, H, Lin, L.-L. | | Deposit date: | 2000-05-24 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway.
Mol.Cell, 5, 2000
|
|
1NY3
 
 | | Crystal structure of ADP bound to MAP KAP kinase 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAP kinase-activated protein kinase 2 | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Maguire, M, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-11 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
1F7T
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE AT 1.8A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1NXK
 
 | | Crystal structure of staurosporine bound to MAP KAP kinase 2 | | Descriptor: | MAP kinase-activated protein kinase 2, STAUROSPORINE, SULFATE ION | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Czerwinski, R.M, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
1F80
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH HOLO-(ACYL CARRIER PROTEIN) | | Descriptor: | ACYL CARRIER PROTEIN, HOLO-(ACYL CARRIER PROTEIN) SYNTHASE, SODIUM ION | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
