6BBN
 
 | |
1WBM
 
 | | HIV-1 protease in complex with symmetric inhibitor, BEA450 | | Descriptor: | (2R,3R,4R,5R)-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]-2,5-BIS(2-PHENYLETHYL)HEXANEDIAMIDE, POL PROTEIN (FRAGMENT) | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease in Complex with Symmetric Inhibitor, Bea450
To be Published
|
|
1FCT
 
 | |
3MBP
 
 | | MALTODEXTRIN-BINDING PROTEIN WITH BOUND MALTOTRIOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
6D34
 
 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
1HFC
 
 | |
4K5A
 
 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Bcl-2-like protein 2, Designed Ankyrin Repeat Protein 013_D12 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
3M6G
 
 | | Crystal structure of actin in complex with lobophorolide | | Descriptor: | (1S,3S,4S,5S,7R,8S,9R,12E,14E,16R,17R,19R)-16-hydroxy-9-{(1S,2S,3S)-2-hydroxy-5-[(2S,4R,6S)-4-methoxy-6-methyltetrahydro-2H-pyran-2-yl]-1,3-dimethylpentyl}-3,5,7,17-tetramethoxy-8,14-dimethyl-11H-spiro[10,23-dioxabicyclo[17.3.1]tricosa-12,14,20-triene-4,2'-oxiran]-11-one, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two molecules of lobophorolide cooperate to stabilize an actin dimer using both their "ring" and "tail" region.
Chem.Biol., 17, 2010
|
|
6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1T4U
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | 2-METHANESULFONYL-BENZENESULFONIC ACID 3-METHYL-5-((1-AMIDINOAMINOOXYMETHYL-CYCLOPROPYL)METHYLOXY)-PHENYLESTER, Hirudin IIIA, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
4K5C
 
 | |
4K5B
 
 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Apoptosis regulator BCL-W, Bcl-2-like protein 2 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
1T4V
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | Hirudin IIIA, N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
2ASM
 
 | | Structure of Rabbit Actin In Complex With Reidispongiolide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2ASP
 
 | | Structure of Rabbit Actin In Complex With Reidispongiolide C | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3MP9
 
 | | Structure of Streptococcal protein G B1 domain at pH 3.0 | | Descriptor: | FORMIC ACID, Immunoglobulin G-binding protein G | | Authors: | Tomlinson, J.H, Green, V.L, Baker, P.J, Williamson, M.P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural origins of pH-dependent chemical shifts in the B1 domain of protein G.
Proteins, 78, 2010
|
|
1G35
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE IN COMPLEX WITH INHIBITOR, AHA024 | | Descriptor: | 2-[4-(HYDROXY-METHOXY-METHYL)-BENZYL]-7-(4-HYDROXYMETHYL-BENZYL)-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1LAMBDA6-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
7K7L
 
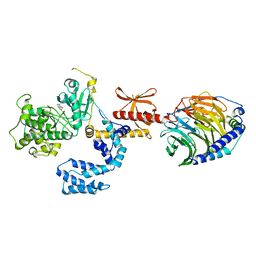 | |
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
4H1G
 
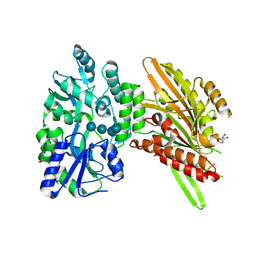 | | Structure of Candida albicans Kar3 motor domain fused to maltose-binding protein | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.A, Duan, D, Delorme, C, Joshi, M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Biochem.Biophys.Res.Commun., 428, 2012
|
|
1YV3
 
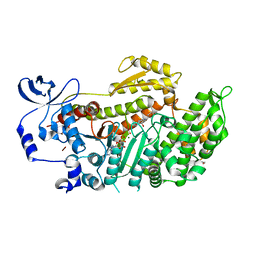 | | The structural basis of blebbistatin inhibition and specificity for myosin II | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Smith, R, Rayment, I. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of blebbistatin inhibition and specificity for myosin II.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7K7Z
 
 | |
1HWQ
 
 | |
3C27
 
 | | Cyanofluorophenylacetamides as Orally Efficacious Thrombin Inhibitors | | Descriptor: | Hirudin-3A, N-[2-(carbamimidamidooxy)ethyl]-2-{6-cyano-3-[(2,2-difluoro-2-pyridin-2-ylethyl)amino]-2-fluorophenyl}acetamide, Thrombin heavy chain, ... | | Authors: | Spurlino, J.C, McMillan, M, Lewandowski, F, Milligan, C. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Cyanofluorophenylacetamides as Orally Efficacious Thrombin Inhibitors
To be Published
|
|
1Z2T
 
 | | NMR structure study of anchor peptide Ser65-Leu87 of enzyme acholeplasma laidlawii Monoglycosyldiacyl Glycerol Synthase (alMGS) in DHPC micelles | | Descriptor: | Anchor peptide Ser65-Leu87 of alMGS | | Authors: | Lind, J, Barany-Wallje, E, Ramo, T, Wieslander, A, Maler, L. | | Deposit date: | 2005-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, position of and membrane-interaction of a putative membrane-anchoring domain of alMGS
To be Published
|
|
