4W4T
 
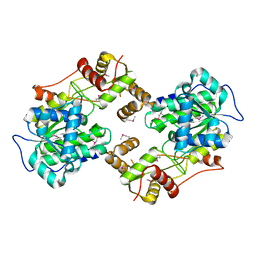 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | 分子名称: | ACETATE ION, MxaA | | 著者 | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | 登録日 | 2014-08-15 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | 分子名称: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | 著者 | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | 登録日 | 2019-04-24 | | 公開日 | 2019-09-11 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
3BZF
 
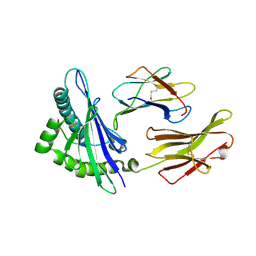 | | The human non-classical major histocompatibility complex molecule HLA-E | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | 著者 | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | 登録日 | 2008-01-17 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
8EZR
 
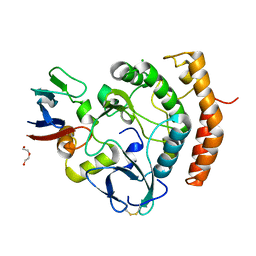 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-11-01 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
3BZE
 
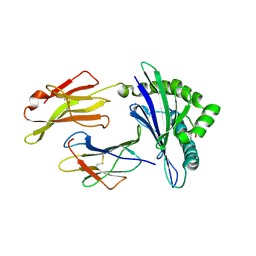 | | The human non-classical major histocompatibility complex molecule HLA-E | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | 著者 | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | 登録日 | 2008-01-17 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
4PJS
 
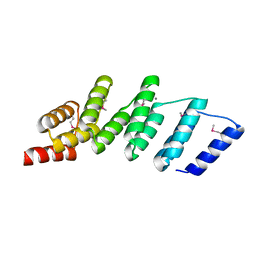 | | Crystal structure of designed (SeMet)-cPPR-NRE protein | | 分子名称: | CALCIUM ION, Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJQ
 
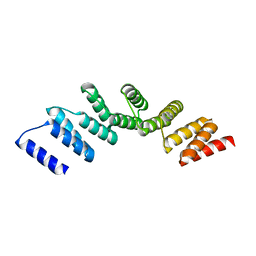 | | Crystal structure of designed cPPR-polyG protein | | 分子名称: | Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.353 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJR
 
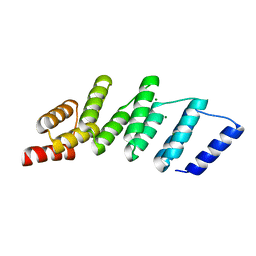 | | Crystal structure of designed cPPR-NRE protein | | 分子名称: | MAGNESIUM ION, Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-05-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4H3B
 
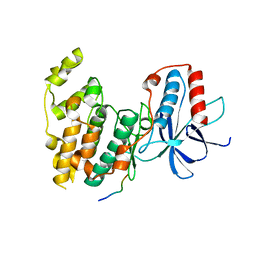 | | Crystal Structure of JNK3 in Complex with SAB Peptide | | 分子名称: | Mitogen-activated protein kinase 10, SH3 domain-binding protein 5 | | 著者 | Nwachukwu, J.C, Laughlin, J.D, Figuera-Losada, M, Cherry, L, Nettles, K.W, LoGrasso, P.V. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural Mechanisms of Allostery and Autoinhibition in JNK Family Kinases.
Structure, 20, 2012
|
|
7TS0
 
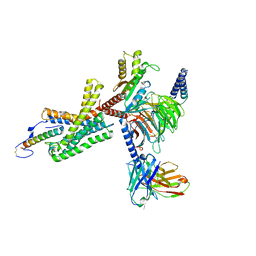 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric Go protein | | 分子名称: | Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Human corticotropin releasing factor receptor 2, Dominant negative Go alpha subunit, G protein gamma subunit, ... | | 著者 | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | 登録日 | 2022-01-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
7TRY
 
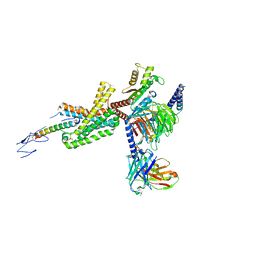 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric G11 protein | | 分子名称: | Corticotropin-releasing factor receptor 2, G protein gamma subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | 登録日 | 2022-01-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
5SY4
 
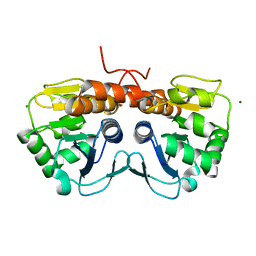 | |
5SY9
 
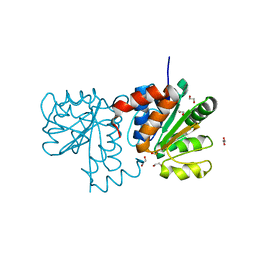 | |
5SY6
 
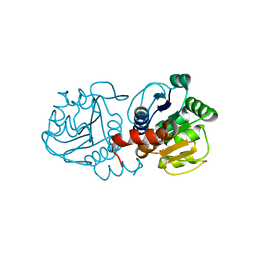 | |
5SYA
 
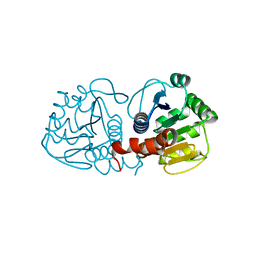 | |
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-18 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-18 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-19 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | 分子名称: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | 著者 | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | 登録日 | 2019-02-19 | | 公開日 | 2020-03-18 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
4IQJ
 
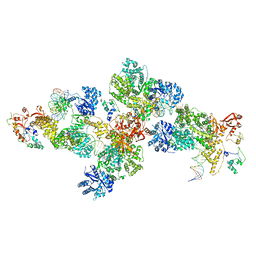 | | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*GP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*CP*GP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*(DOC))-3'), ... | | 著者 | Liu, B, Lin, J, Steitz, T. | | 登録日 | 2013-01-11 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of PolIIIalpha-Tauc-DNA complex suggests an atomic model of the replisome
Structure, 21, 2013
|
|
6W20
 
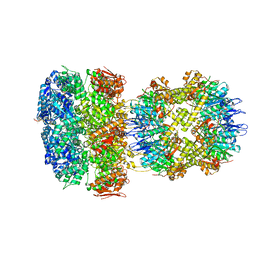 | | ClpAP Disengaged State bound to RepA-GFP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | 著者 | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4JR5
 
 | | Structure-based Identification of Ureas as Novel Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-(piperidin-1-ylsulfonyl)phenyl]-3-(pyridin-3-ylmethyl)thiourea, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Gunzner-Toste, J, Zhao, G, Yuen, P, Bair, K.W. | | 登録日 | 2013-03-21 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
6W21
 
 | | ClpAP Engaged2 State bound to RepA-GFP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | 著者 | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4JNM
 
 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | 分子名称: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | 登録日 | 2013-03-15 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6W1Z
 
 | | ClpAP Engaged1 State bound to RepA-GFP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | 著者 | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
