3I9N
 
 | | Crystal structure of human CD38 complexed with an analog ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
6JZ5
 
 | |
6JZ3
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin | | Descriptor: | (2~{S},3~{R},4~{R},5~{S})-3,4,5-tris(oxidanyl)piperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
3I9K
 
 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9M
 
 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
6IS9
 
 | | Crystal Structure of ZmMOC1 | | Descriptor: | Monokaryotic chloroplast 1 | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JZ2
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ4
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ7
 
 | |
3I9J
 
 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9L
 
 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
6JZ6
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ1
 
 | |
5YWH
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with Y13508 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION, [1,3-diethyl-2,2-bis(oxidanylidene)-2$l^{6},1,3-benzothiadiazol-5-yl]-(1-methyl-5-oxidanyl-pyrazol-4-yl)methanone | | Authors: | Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Arabidopsis thaliana HPPD complexed with Y13508
To be published
|
|
5YWK
 
 | | Crystal structure of Arabidopsis thaliana HPPD complexed with Benquitrione-Methyl | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Lin, H.Y. | | Deposit date: | 2017-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure-Guided Discovery of Silicon-Containing Subnanomolar Inhibitor of Hydroxyphenylpyruvate Dioxygenase as a Potential Herbicide.
J.Agric.Food Chem., 69, 2021
|
|
4V98
 
 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
8HZA
 
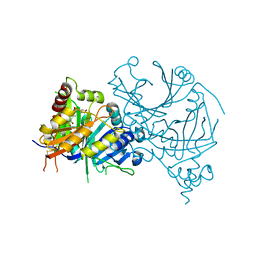 | | Crystal structure of AtHPPD-YH20326 complex | | Descriptor: | 1,4-dimethyl-3-(oxan-4-ylmethyl)-5-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-benzimidazol-2-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of AtHPPD-YH20326 complex
To Be Published
|
|
8HXH
 
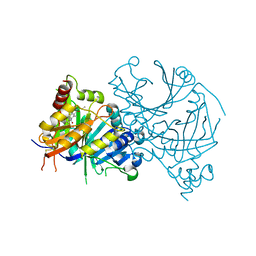 | | Crystal structure of AtHPPD-Y18024 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-propyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystal structure of AtHPPD-Y18024 complex
To Be Published
|
|
8HYS
 
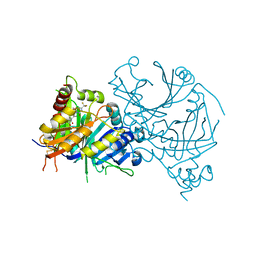 | | Crystal structure of AtHPPD-Y19802 complex | | Descriptor: | 1-methyl-3-[2-methyl-6-methylsulfonyl-3-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-phenyl]-6-(trifluoromethyl)pyrimidine-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of AtHPPD-Y19802 complex
To Be Published
|
|
8HZ7
 
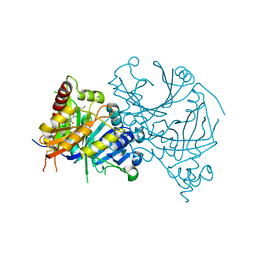 | | Crystal structure of AtHPPD-Y181135 complex | | Descriptor: | 3-butyl-5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-1,2,3-benzotriazin-4-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-08 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of AtHPPD-Y181135 complex
To Be Published
|
|
8I0G
 
 | | Crystal structure of AtHPPD-Y19543 complex | | Descriptor: | 3-[(4-fluorophenyl)methyl]-5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-1,2,3-benzotriazin-4-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystal structure of AtHPPD-Y19543 complex
To Be Published
|
|
8HWR
 
 | | Crystal structure of AtHPPD-Y191193 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-chloranyl-3-cyclopentyl-1-methyl-7-(2-oxidanyl-6-oxidanylidene-cyclohexa-1,3-dien-1-yl)carbonyl-quinazoline-2,4-dione, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of AtHPPD-Y191193 complex
To Be Published
|
|
8I2P
 
 | | Crystal structure of AtHPPD-Y19060 complex | | Descriptor: | 3-(cyclopropylmethyl)-6-[(5-cyclopropyl-2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-14 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of AtHPPD-Y19060 complex
To Be Published
|
|
8HYP
 
 | | Crystal structure of AtHPPD-Y18400 complex | | Descriptor: | 3-[(4-fluorophenyl)methyl]-1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Crystal structure of AtHPPD-Y18400 complex
To Be Published
|
|
8I36
 
 | | Crystal structure of AtHPPD-Y18980 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-1-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Crystal structure of AtHPPD-Y18980 complex
To Be Published
|
|
