4BX2
 
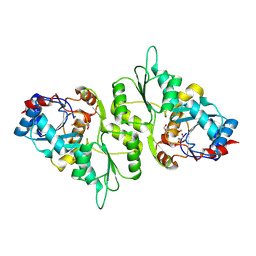 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in complex with Beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop.
J.Biol.Chem., 289, 2014
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2000-04-13 | | Release date: | 2000-08-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
4EYI
 
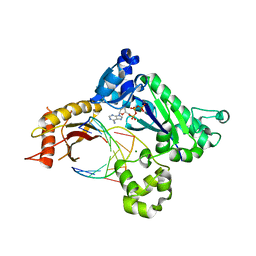 | |
3ESW
 
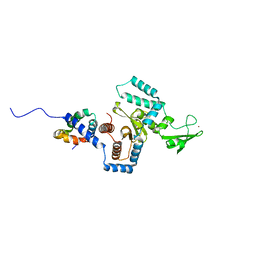 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
2HPL
 
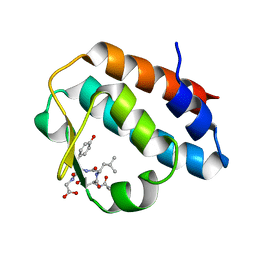 | | Crystal structure of the mouse p97/PNGase complex | | Descriptor: | C-terminal of mouse p97/VCP, PNGase | | Authors: | Zhao, G, Zhou, X, Wang, L, Li, G, Lennarz, W, Schindelin, H. | | Deposit date: | 2006-07-17 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4BX3
 
 | |
3PR4
 
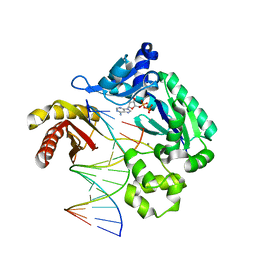 | | Dpo4 Y12A mutant incorporating dADP opposite template dT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Kirouac, K.N, Suo, Z, Ling, H. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural mechanism of ribonucleotide discrimination by a Y-family DNA polymerase.
J.Mol.Biol., 407, 2011
|
|
3Q8S
 
 | |
2FN9
 
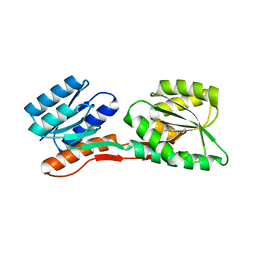 | | Thermotoga maritima Ribose Binding Protein Unliganded Form | | Descriptor: | ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Tian, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-01-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in a thermophilic ribose-binding protein.
Bmc Struct.Biol., 8, 2008
|
|
2RDJ
 
 | |
3CMM
 
 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
2FTS
 
 | |
4F9Z
 
 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
2IPN
 
 | |
2IPM
 
 | |
4FBT
 
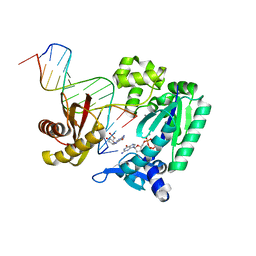 | |
1CFF
 
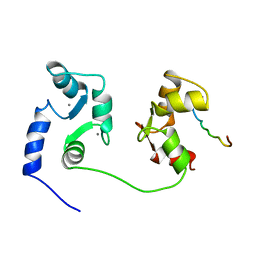 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
3QEA
 
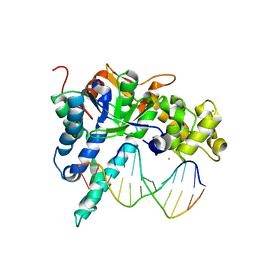 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with DNA (complex II) | | Descriptor: | BARIUM ION, DNA (5'-D(P*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Orans, J, McSweeney, E.A, Iyer, R.R, Hast, M.A, Hellinga, H.W, Modrich, P, Beese, L.S. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QQ7
 
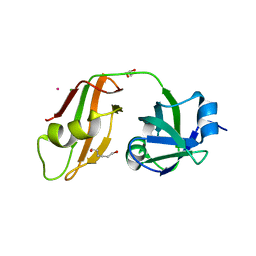 | |
2IOY
 
 | |
2G9G
 
 | | Crystal structure of His-tagged mouse PNGase C-terminal domain | | Descriptor: | GLYCEROL, SULFATE ION, peptide N-glycanase | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4FBU
 
 | | Dpo4 polymerase pre-insertion binary complex with the N-(deoxyguanosin-8-yl)-1-aminopyrene lesion | | Descriptor: | CALCIUM ION, DNA polymerase IV, DNA primer, ... | | Authors: | Kirouac, K, Basu, A, Ling, H. | | Deposit date: | 2012-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of replication stalling on a bulky amino-polycyclic aromatic hydrocarbon DNA adduct by a y family DNA polymerase.
J.Mol.Biol., 425, 2013
|
|
2FU3
 
 | |
2IPL
 
 | |
2NQS
 
 | | MoeA E188A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
