3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
1TV8
 
 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3HP4
 
 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2GD5
 
 | | Structural basis for budding by the ESCRTIII factor CHMP3 | | Descriptor: | Charged multivesicular body protein 3 | | Authors: | Muziol, T.M, Pineda-Molina, E, Ravelli, R.B, Zamborlini, A, Usami, Y, Gottlinger, H, Weissenhorn, W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Budding by the ESCRT-III Factor CHMP3.
Dev.Cell, 10, 2006
|
|
5W2A
 
 | |
3GAE
 
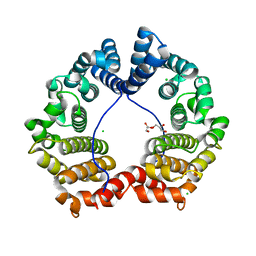 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5W2C
 
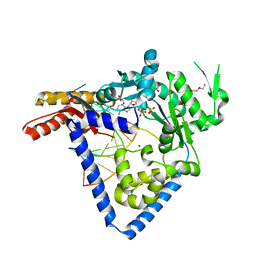 | |
3GV5
 
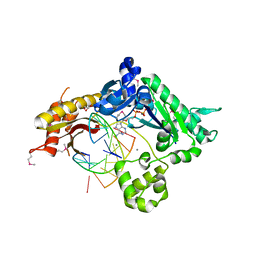 | |
3I5O
 
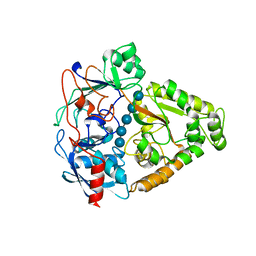 | | The X-ray crystal structure of a thermophilic cellobiose binding protein bound with cellopentaose | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cuneo, M.J, Hellinga, H.W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Semi-specific Oligosaccharide Recognition by a Cellulose-binding Protein of Thermotoga maritima Reveals Adaptations for Functional Diversification of the Oligopeptide Periplasmic Binding Protein Fold.
J.Biol.Chem., 284, 2009
|
|
1TV7
 
 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6BS1
 
 | |
1JW9
 
 | | Structure of the Native MoeB-MoaD Protein Complex | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
6CST
 
 | | Structure of human DNA polymerase kappa with DNA | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Jha, V, Ling, H. | | Deposit date: | 2018-03-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 angstrom resolution crystal structure of human pol kappa reveals a new catalytic function of N-clasp in DNA replication.
Sci Rep, 8, 2018
|
|
1JIZ
 
 | | Crystal Structure Analysis of human Macrophage Elastase MMP-12 | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, ZINC ION, ... | | Authors: | Nar, H, Werle, K, Bauer, M.M.T, Dollinger, H, Jung, B. | | Deposit date: | 2001-07-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human macrophage elastase (MMP-12) in complex with a hydroxamic acid inhibitor.
J.Mol.Biol., 312, 2001
|
|
2IO4
 
 | | Crystal structure of PCNA12 dimer from Sulfolobus solfataricus. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DNA polymerase sliding clamp B, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-10-09 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6BRX
 
 | |
1JLJ
 
 | | 1.6 Angstrom crystal structure of the human neuroreceptor anchoring and molybdenum cofactor biosynthesis protein gephyrin | | Descriptor: | FORMIC ACID, SODIUM ION, gephyrin | | Authors: | Schwarz, G, Schrader, N, Mendel, R.R, Hecht, H.-J, Schindelin, H. | | Deposit date: | 2001-07-16 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human gephyrin and plant Cnx1 G domains: comparative analysis and functional implications.
J.Mol.Biol., 312, 2001
|
|
2NTI
 
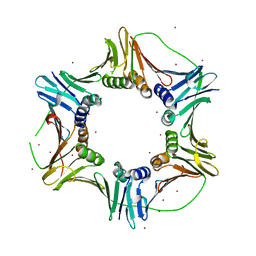 | | Crystal structure of PCNA123 heterotrimer. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, BROMIDE ION, DNA polymerase sliding clamp A, ... | | Authors: | Hlinkova, V, Ling, H. | | Deposit date: | 2006-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2IJX
 
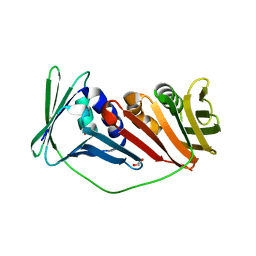 | |
1K6Y
 
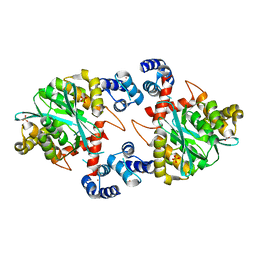 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
1ZRY
 
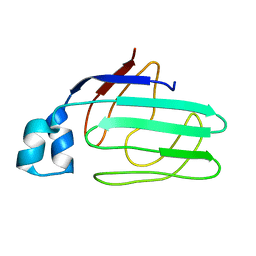 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
8TG1
 
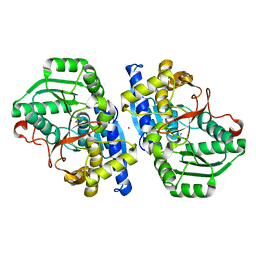 | | Caldicellulosiruptor saccharolyticus periplasmic urea-binding protein | | Descriptor: | BROMIDE ION, Extracellular ligand-binding receptor, UREA | | Authors: | Allert, M.J, Kumar, S, Wang, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure-based functional analysis reveals multiple roles and widespread use of urea-binding proteins in nitrogen metabolism
To Be Published
|
|
7ZH9
 
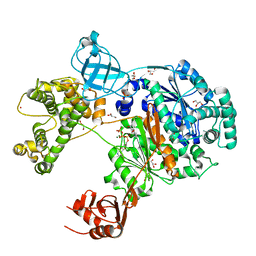 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7ZTL
 
 | |
8A1I
 
 | |
