8HFD
 
 | | Crystal structure of allantoinase from E. coli BL21 | | Descriptor: | Allantoinase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Lin, E.S, Huang, H.Y, Yang, P.C, Liu, H.W, Huang, C.Y. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis.
Molecules, 28, 2023
|
|
2Q8K
 
 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
7VUM
 
 | | Crystal structure of SSB complexed with que | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity.
Int J Mol Sci, 23, 2022
|
|
7DEP
 
 | | S. aureus SsbB with 5-FU | | Descriptor: | 5-FLUOROURACIL, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.094 Å) | | Cite: | Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein
Biochem.Biophys.Res.Commun., 534, 2020
|
|
7D8J
 
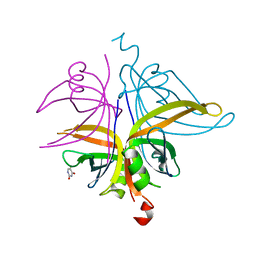 | | S. aureus SsbB with 5-FU | | Descriptor: | 5-FLUOROURACIL, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2020-10-08 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein
Biochem.Biophys.Res.Commun., 534, 2020
|
|
7F2N
 
 | |
5JEA
 
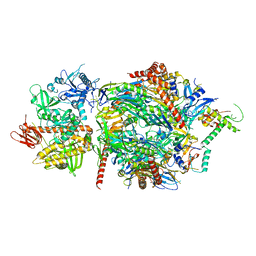 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
8IM5
 
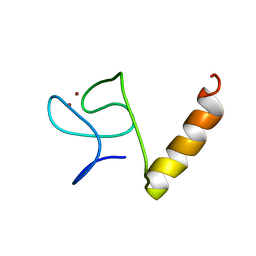 | |
4ZKE
 
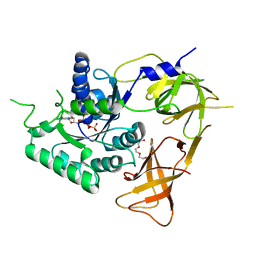 | |
4ZKD
 
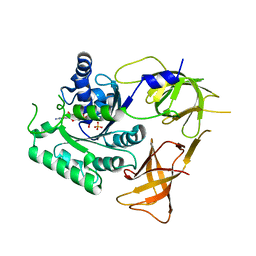 | |
2MGW
 
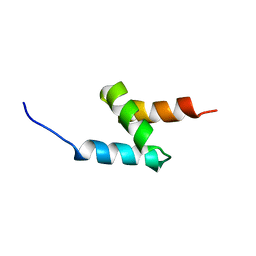 | | Solution Structure of the UBA Domain of Human NBR1 | | Descriptor: | Next to BRCA1 gene 1 protein | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-11-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
2MJ5
 
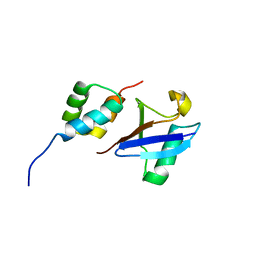 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | Descriptor: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-12-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
4A2P
 
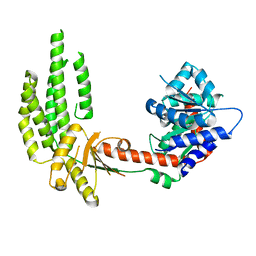 | | Structure of duck RIG-I helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4AVQ
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AWF
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with 2 4-dioxo-4-phenylbutanoic acid DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4A2X
 
 | | Structure of duck RIG-I C-terminal domain (CTD) with 14-mer dSRNA | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*UP*GP*UP*UP*CP*UP*CP*CP*CP)-3', 5'-R(*GP*GP*GP*AP*GP*AP*AP*CP*AP*AP*CP*GP*CP*GP)-3', RETINOIC ACID INDUCIBLE PROTEIN I, ... | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA.
Cell, 147, 2011
|
|
4A36
 
 | | Structure of duck RIG-I helicase domain bound to 19-mer dsRNA and ATP transition state analogue | | Descriptor: | 5'-R(*GP*CP*AP*UP*GP*CP*GP*AP*CP*CP*UP*CP*UP*GP *UP*UP*UP*GP*A)-3', 5'-R(*UP*CP*AP*AP*AP*CP*AP*GP*AP*GP*GP*UP*CP*GP *CP*AP*UP*GP*C)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2V
 
 | | Structure of duck RIG-I C-terminal domain (CTD) | | Descriptor: | GLYCEROL, RETINOIC ACID INDUCIBLE PROTEIN I, ZINC ION | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4AVL
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with dTMP | | Descriptor: | MANGANESE (II) ION, POLYMERASE PA, THYMIDINE-5'-PHOSPHATE | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H.J, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2012-05-28 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AWH
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with rUMP | | Descriptor: | MANGANESE (II) ION, POLYMERASE PA, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AVG
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with diketo compound 2 | | Descriptor: | (Z)-4-[4-[(4-chlorophenyl)methyl]-1-(cyclohexylmethyl)piperidin-4-yl]-2-oxidanyl-4-oxidanylidene-but-2-enoic acid, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-05-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4A2W
 
 | | Structure of full-length duck RIG-I | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
2VVQ
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with the inhibitor 5-deoxy-5-phospho-D- ribonate | | Descriptor: | 5-O-phosphono-D-ribonic acid, RIBOSE-5-PHOSPHATE ISOMERASE B, SULFATE ION | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
4A2Q
 
 | | Structure of duck RIG-I tandem CARDs and helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
2VVP
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
