7YHE
 
 | | Crystal structure of the triple mutant CmnC-L136Q,S138G,D249Y in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, CmnC, FE (III) ION, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Hsiao, P.Y, Chang, C.Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7YW9
 
 | | Crystal structure of the triple mutant CmnC-L136Q,S138G,D249Y in complex with alpha-KG | | Descriptor: | ACETATE ION, CmnC, D-ARGININE, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Hsiao, P.Y, Chang, C.Y. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y0G
 
 | | Crystal structure of anti-mPEG h15-2b Fab | | Descriptor: | 15-2b heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Lin, E.C, Su, Y.C. | | Deposit date: | 2022-06-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group.
Commun Chem, 5, 2022
|
|
7VGN
 
 | | Crystal structure of CmnC | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CmnC, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7VGL
 
 | | Crystal structure of CmnC | | Descriptor: | ACETATE ION, CmnC | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Zheng, Y.Z, Chang, C.Y. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5I
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | ARGININE, CmnC, FE (III) ION, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5P
 
 | | Crystal structure of CmnC in complex with L-arginine and alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, CmnC, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7Y5F
 
 | | Crystal structure of CmnC in complex with L-homoarginine | | Descriptor: | CmnC, FE (III) ION, L(+)-TARTARIC ACID, ... | | Authors: | Hsiao, Y.H, Huang, S.J, Lin, E.C, Lee, Y.C, Chang, C.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
7V5L
 
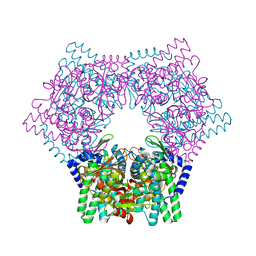 | | Crystal structure of human bleomycin hydrolase | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7V5S
 
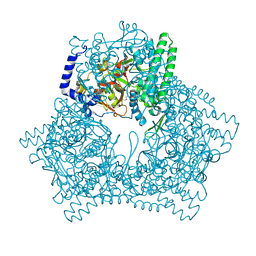 | | Crystal structure of human bleomycin hydrolase C73A mutant | | Descriptor: | Bleomycin hydrolase, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7V5T
 
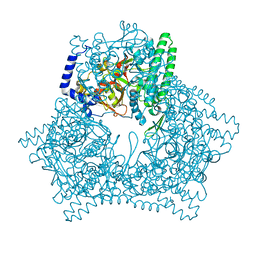 | | Crystal structure of human bleomycin hydrolase C73S mutant | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7XF9
 
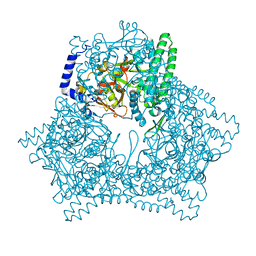 | | Crystal structure of human bleomycin hydrolase H372A mutant | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
2N28
 
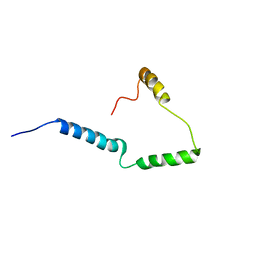 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2N29
 
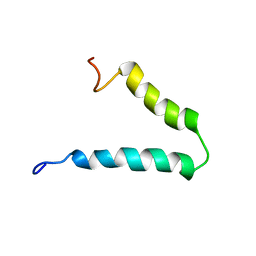 | | Solution-state NMR structure of Vpu cytoplasmic domain | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
6SLZ
 
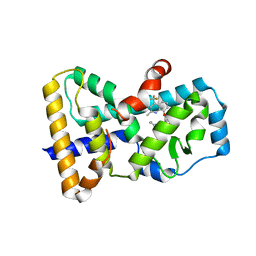 | | Crystal structure of human ROR gamma LBD in complex with a (quinolinoxymethyl)benzamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[2-methyl-4-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]carbonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human ROR gamma LBD in complex with a (quinolinoxymethyl)benzamide inverse agonist
To Be Published
|
|
6Q2W
 
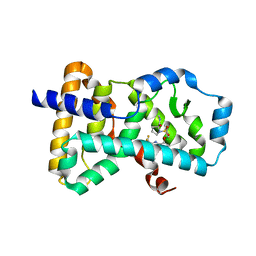 | | Crystal structure of human ROR gamma LBD in complex with a quinoline sulfonamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[4-imidazol-1-yl-2-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]sulfonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Ciesielski, F, Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8TB5
 
 | | TYK2 JH2 bound to Compound7 | | Descriptor: | ACETATE ION, N-{(3P)-3-[3-(dimethylsulfamoyl)phenyl]-1H-pyrrolo[2,3-c]pyridin-5-yl}cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
8TB6
 
 | | TYK2 JH2 bound to Compound14 | | Descriptor: | N-[(3M)-3-{6-[(3R)-3-methoxyoxolan-3-yl]pyridin-2-yl}-1-methyl-1H-pyrrolo[2,3-c]pyridin-5-yl]urea, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
