6SX7
 
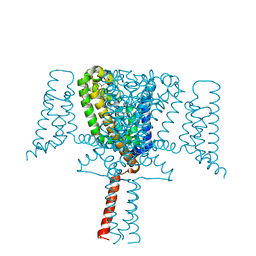 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) (2.2 Angstrom resolution) | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SXG
 
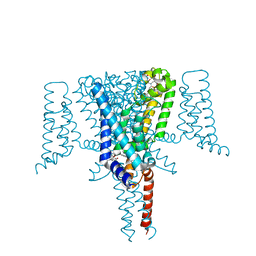 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs in complex with 4-hydroxytamoxifen (2.4 Angstrom resolution) | | Descriptor: | 4-HYDROXYTAMOXIFEN, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
1XG2
 
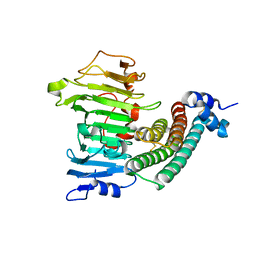 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
8PK3
 
 | | CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer molecule (A31), ... | | Authors: | Debski-Antoniak, O, Flynn, A, Klebl, D.P, Tiede, C, Muench, S, Tomlinson, D, Fontana, J. | | Deposit date: | 2023-06-24 | | Release date: | 2024-01-03 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting the Affimer platform against influenza A virus.
Mbio, 15, 2024
|
|
8Q9S
 
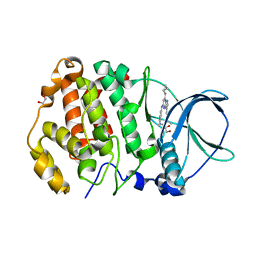 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE INHIBITOR SGC-CK2-1 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha', ~{N}-[5-[[3-cyano-7-(cyclopropylamino)-3~{H}-pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-methyl-phenyl]propanamide | | Authors: | Werner, C, Lindenblatt, D, Niefind, K. | | Deposit date: | 2023-08-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | Discovery and Exploration of Protein Kinase CK2 Binding Sites Using CK2alpha Cys336Ser as an Exquisite Crystallographic Tool
Kinases Phosphatases, 2023
|
|
8RTZ
 
 | | The structure of E. coli penicillin binding protein 3 (PBP3) in complex with a bicyclic peptide inhibitor | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Bicyclic peptide inhibitor, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Rowland, C.E, Dods, R, Lewis, N, Stanway, S.J, Bellini, D, Beswick, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and chemical optimisation of a Potent, Bi-cyclic (Bicycle) Antimicrobial Inhibitor of Escherichia coli PBP3
To Be Published
|
|
9FCR
 
 | |
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
8A8F
 
 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
5FR7
 
 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
5FRK
 
 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-18 | | Release date: | 2017-02-15 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
3TE5
 
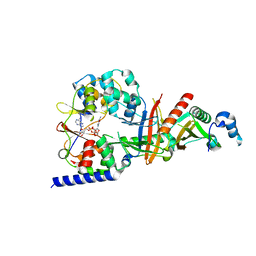 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
7LNM
 
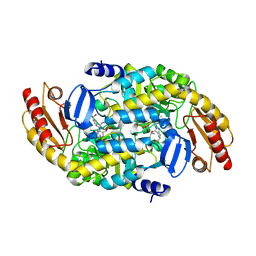 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
3T4N
 
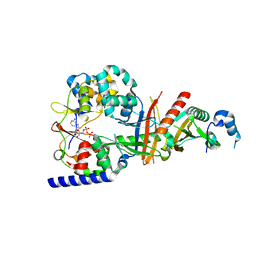 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TDH
 
 | | Structure of the regulatory fragment of sccharomyces cerevisiae AMPK in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
2Y8Q
 
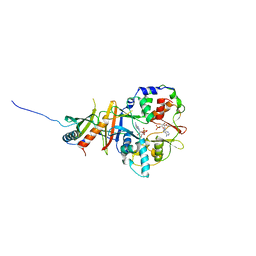 | | Structure of the regulatory fragment of mammalian AMPK in complex with one ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2Y8L
 
 | | Structure of the regulatory fragment of mammalian aMPK in complex with two ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2YA3
 
 | | STRUCTURE OF THE REGULATORY FRAGMENT OF MAMMALIAN AMPK IN COMPLEX WITH COUMARIN ADP | | Descriptor: | 3'-(7-diethylaminocoumarin-3-carbonylamino)-3'-deoxy-ADP, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
7BAG
 
 | | C3b in complex with CP40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, AMINO GROUP, ... | | Authors: | Lamers, C, Xue, X, Smiesko, M, van Son, H, Wagner, B, Sfyroera, G, Gros, P, Lambris, J, Ricklin, D. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into mode-of-action and structural determinants of the compstatin family of clinical complement inhibitors.
Nat Commun, 13, 2022
|
|
2WPA
 
 | | Optimisation of 6,6-Dimethyl Pyrrolo 3,4-c pyrazoles: Identification of PHA-793887, a Potent CDK Inhibitor Suitable for Intravenous Dosing | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-{6,6-DIMETHYL-5-[(1-METHYLPIPERIDIN-4-YL)CARBONYL]-1,4,5,6-TETRAHYDROPYRROLO[3,4-C]PYRAZOL-3-YL}-3-METHYLBUTANAMIDE, ... | | Authors: | Brasca, M.G, Albanese, C, Alzani, R, Amici, R, Avanzi, N, Ballinari, D, Bischoff, J, Borghi, D, Casale, E, Croci, V, Fiorentini, F, Isacchi, A, Mercurio, C, Nesi, M, Orsini, P, Pastori, W, Pesenti, E, Pevarello, P, Roussel, P, Varasi, M, Volpi, D, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Optimization of 6,6-Dimethyl Pyrrolo[3,4-C]Pyrazoles: Identification of Pha-793887, a Potent Cdk Inhibitor Suitable for Intravenous Dosing.
Bioorg.Med.Chem., 18, 2010
|
|
