2NVF
 
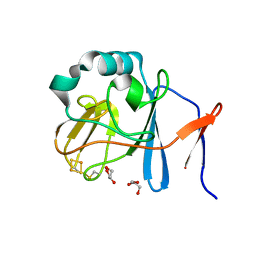 | | Soluble domain of Rieske Iron-Sulfur protein. | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-12 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NWF
 
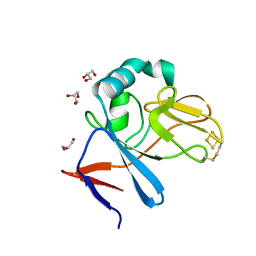 | | Soluble domain of Rieske Iron Sulfur Protein | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-14 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVG
 
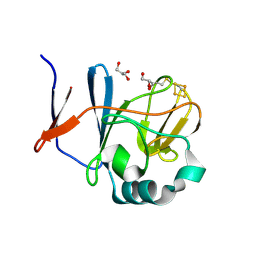 | | Soluble domain of Rieske Iron Sulfur protein. | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-12 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2KCR
 
 | |
2JQW
 
 | |
2JOM
 
 | |
2KLZ
 
 | |
6Y7M
 
 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Hilgenfeld, R. | | 登録日 | 2020-03-01 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6ZZR
 
 | | The Crystal Structure of human LDHA from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-lactate dehydrogenase A chain | | 著者 | Wang, F, Lin, D, Cheng, W, Bao, X, Zhu, B, Shang, H. | | 登録日 | 2020-08-05 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The Crystal Structure of human LDHA from Biortus
To Be Published
|
|
6X9H
 
 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | 分子名称: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | 著者 | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | 登録日 | 2020-06-02 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
6Y2G
 
 | | Crystal structure (orthorhombic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | 分子名称: | Green fluorescent protein | | 著者 | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | 登録日 | 2023-04-26 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.537 Å) | | 主引用文献 | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
3E9F
 
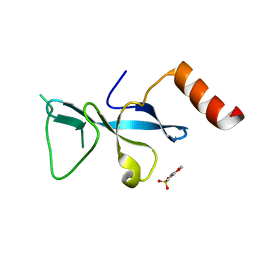 | | Crystal structure short-form (residue1-113) of Eaf3 chromo domain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromatin modification-related protein EAF3 | | 著者 | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | 登録日 | 2008-08-22 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3E9G
 
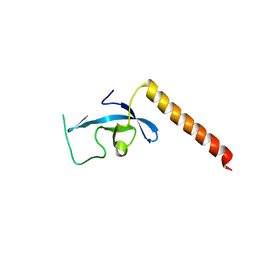 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | 分子名称: | Chromatin modification-related protein EAF3 | | 著者 | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | 登録日 | 2008-08-22 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
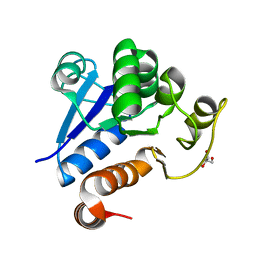
7C62
 
 | |
7CMR
 
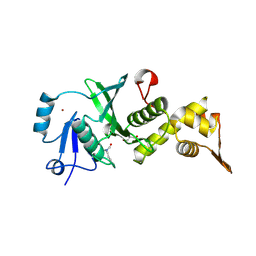 | | The Crystal Structure of human MYST1 from Biortus. | | 分子名称: | GLYCEROL, Histone acetyltransferase KAT8, ZINC ION | | 著者 | Wang, F, Lin, D, Lv, Z, Xu, X, Tan, J, Shang, H. | | 登録日 | 2020-07-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Crystal Structure of human MYST1 from Biortus.
To Be Published
|
|
7CM2
 
 | | The Crystal Structure of human USP7 USP domain from Biortus | | 分子名称: | GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Wang, F, Cheng, W, Lv, Z, Lin, D, Zhu, B, Miao, Q, Bao, X, Shang, H. | | 登録日 | 2020-07-24 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The Crystal Structure of human USP7 USP domain from Biortus.
To Be Published
|
|
7CML
 
 | | The Crystal Structure of human JNK2 from Biortus. | | 分子名称: | Mitogen-activated protein kinase 9 | | 著者 | Wang, F, Lin, D, Cheng, W, Miao, Q, Huang, Y, Shang, H. | | 登録日 | 2020-07-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Crystal Structure of human JNK2 from Biortus.
To Be Published
|
|
7CVP
 
 | | The Crystal Structure of human PHGDH from Biortus. | | 分子名称: | D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Wang, F, Lv, Z, Cheng, W, Lin, D, Miao, Q, Huang, Y. | | 登録日 | 2020-08-26 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Crystal Structure of human PHGDH from Biortus.
To Be Published
|
|
7C4I
 
 | |
7CA4
 
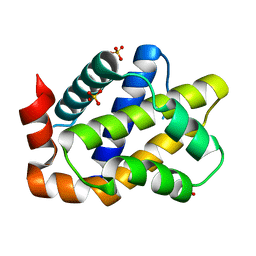 | |
7DSF
 
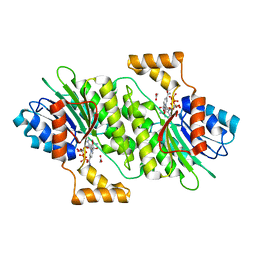 | | The Crystal Structure of human SPR from Biortus. | | 分子名称: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | 著者 | Wang, F, Lv, Z, Cheng, W, Lin, D, Meng, Q, Zhang, B, Huang, Y. | | 登録日 | 2020-12-31 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Crystal Structure of human SPR from Biortus.
To Be Published
|
|
7DS7
 
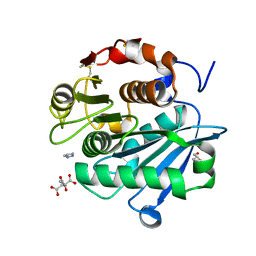 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | 分子名称: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | 著者 | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | 登録日 | 2020-12-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
7D2C
 
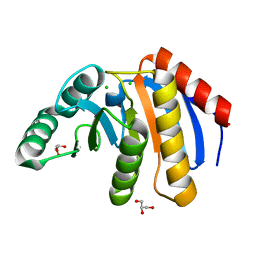 | | The Crystal Structure of human PARP14 from Biortus. | | 分子名称: | CHLORIDE ION, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP14 | | 著者 | Wang, F, Miao, Q, Lv, Z, Cheng, W, Lin, D, Xu, X, Tan, J. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | The Crystal Structure of human PARP14 from Biortus.
To Be Published
|
|
