2OPU
 
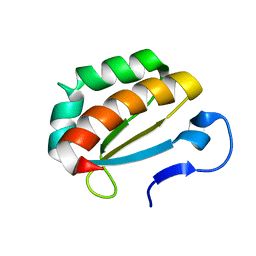 | |
3TE5
 
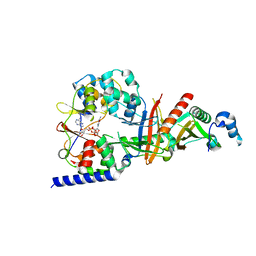 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
4B8T
 
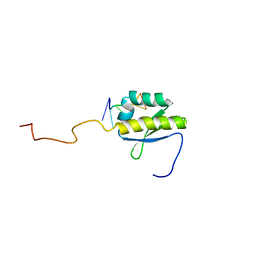 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
2QFA
 
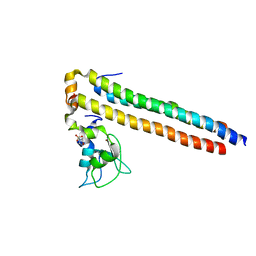 | | Crystal structure of a Survivin-Borealin-INCENP core complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Baculoviral IAP repeat-containing protein 5, Borealin, ... | | Authors: | Jeyaprakash, A.A, Klein, U.R, Lindner, D, Ebert, J, Nigg, E.A, Conti, E. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Survivin-Borealin-INCENP Core Complex Reveals How Chromosomal Passengers Travel Together.
Cell(Cambridge,Mass.), 131, 2007
|
|
4CFF
 
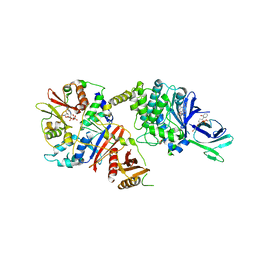 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
2BKD
 
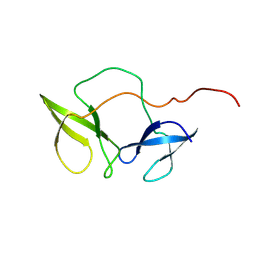 | | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | | Descriptor: | Fragile X messenger ribonucleoprotein 1 | | Authors: | Ramos, A, Hollingworth, D, Adinolfi, S, Castets, M, Kelly, G, Frenkiel, T.A, Bardoni, B, Pastore, A. | | Deposit date: | 2005-02-15 | | Release date: | 2006-01-18 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction.
Structure, 14, 2006
|
|
7JW9
 
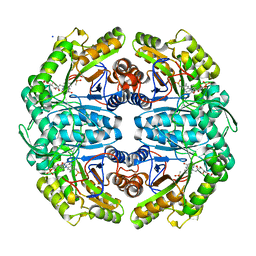 | | Ternary cocrystal structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, SODIUM ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7K64
 
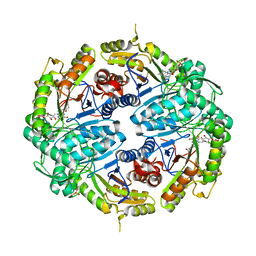 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JYB
 
 | | Binary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JTJ
 
 | |
4CFE
 
 | | Structure of full length human AMPK in complex with a small molecule activator, a benzimidazole derivative (991) | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3LEO
 
 | | Structure of human Leukotriene C4 synthase mutant R31Q in complex with glutathione | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Niegowski, D, Martinez-Molina, D, Rinaldo-Matthis, A, Nordlund, P, Haeggstrom, J. | | Deposit date: | 2010-01-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 104 is a key catalytic residue in leukotriene C4 synthase.
J.Biol.Chem., 285, 2010
|
|
2MVO
 
 | | Solution structure of the lantibiotic self-resistance lipoprotein MlbQ from Microbispora ATCC PTA-5024 | | Descriptor: | Putative lipoprotein | | Authors: | Pozzi, R, Schwartz, P, Linke, D, Kulik, A, Nega, M, Wohlleben, W, Stegmann, E, Coles, M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024.
Environ Microbiol, 18, 2016
|
|
2Y8Q
 
 | | Structure of the regulatory fragment of mammalian AMPK in complex with one ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2Y8L
 
 | | Structure of the regulatory fragment of mammalian aMPK in complex with two ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2YA3
 
 | | STRUCTURE OF THE REGULATORY FRAGMENT OF MAMMALIAN AMPK IN COMPLEX WITH COUMARIN ADP | | Descriptor: | 3'-(7-diethylaminocoumarin-3-carbonylamino)-3'-deoxy-ADP, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2N8L
 
 | |
2N8M
 
 | |
4AZ3
 
 | | crystal structure of cathepsin a, complexed with 15a | | Descriptor: | (3S)-3-({[1-(2-fluorophenyl)-5-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-1H-pyrazol-3-yl]carbonyl}amino)-3-(2-methylphenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ruf, S, Buning, C, Schreuder, H, Horstick, G, Linz, W, Olpp, T, Pernerstorfer, J, Hiss, K, Kroll, K, Kannt, A, Kohlmann, M, Linz, D, Huebschle, T, Ruetten, H, Wirth, K, Schmidt, T, Sadowski, T. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Novel Beta-Amino Acid Derivatives as Inhibitors of Cathepsin A.
J.Med.Chem., 55, 2012
|
|
7JUA
 
 | |
8OL6
 
 | | Murine type II Abeta fibril from tgAPPSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL2
 
 | | Murine type II Abeta fibril from APP23 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL5
 
 | | Murine type II Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
