4DJ1
 
 | |
3DVQ
 
 | |
3DW1
 
 | |
3IJU
 
 | |
3DZN
 
 | |
3E0A
 
 | |
1AFE
 
 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
4MKN
 
 | | Crystal structure of chloroplastic triosephosphate isomerase from Chlamydomonas reinhardtii at 1.1 A of resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Triosephosphate isomerase | | Authors: | Fermani, S, Sciabolini, C, Zaffagnini, M, Lemaire, S.D. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structure and Redox Properties of Chloroplastic Triosephosphate Isomerase from Chlamydomonas reinhardtii.
Mol Plant, 7, 2014
|
|
3I1H
 
 | | Crystal structure of human BFL-1 in complex with BAK BH3 peptide | | Descriptor: | Apoptosis regulator BAK, Protein BFL-1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T, White, E, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of human BFL-1 in complex with BAK BH3 peptide
To be Published
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
3DE3
 
 | |
3DE5
 
 | |
3DO1
 
 | |
3DO0
 
 | |
4OXN
 
 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OIM
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
3IJV
 
 | |
4Q2Z
 
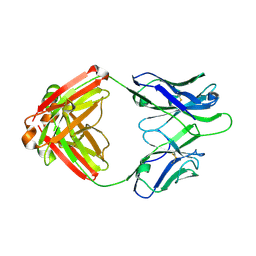 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | Descriptor: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | Authors: | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | Deposit date: | 2014-04-10 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
2WJ5
 
 | | Rat alpha crystallin domain | | Descriptor: | HEAT SHOCK PROTEIN BETA-6 | | Authors: | Naylor, C.E, Bagneris, C, Bateman, O.A, Cronin, N, Keep, N.H, Slingsby, C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structures of Alpha-Crystallin Domain Dimers of Alphab-Crystallin and Hsp20.
J.Mol.Biol., 392, 2009
|
|
1I0A
 
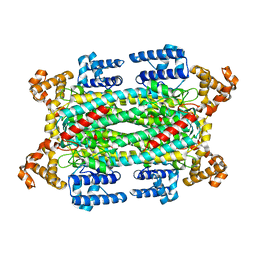 | |
1HY1
 
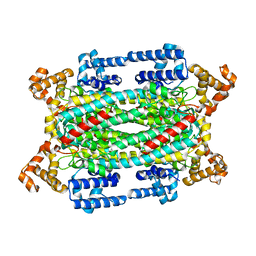 | |
2A52
 
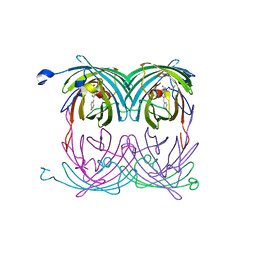 | | fluorescent protein asFP595, S158V, on-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A50
 
 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A53
 
 | | fluorescent protein asFP595, A143S, off-state | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1HK0
 
 | | Human GammaD Crystallin Structure at 1.25 A Resolution | | Descriptor: | Gamma-crystallin D | | Authors: | Basak, A.K, Slingsby, C. | | Deposit date: | 2003-03-05 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-Resolution X-Ray Crystal Structures of Human Gammad Crystallin (1.25A) and the R58H Mutant (1.15A) Associated with Aculeiform Cataract
J.Mol.Biol., 328, 2003
|
|
