6EEH
 
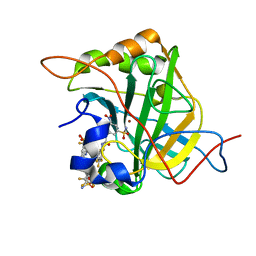 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-14 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EEO
 
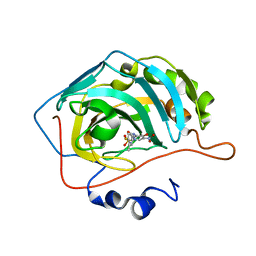 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 3-{[(4-fluoro-3-methylphenyl)carbamoyl]amino}-4-hydroxy-5-nitrobenzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6EDA
 
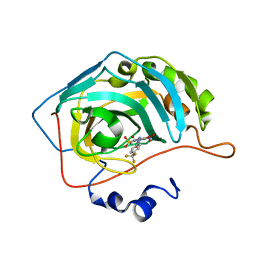 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-09 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6ECZ
 
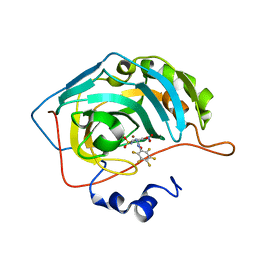 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
3EGA
 
 | |
3EGB
 
 | |
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WQH
 
 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WLT
 
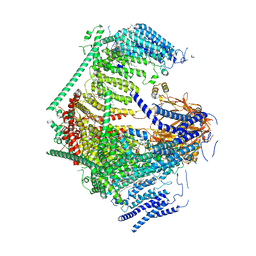 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WLU
 
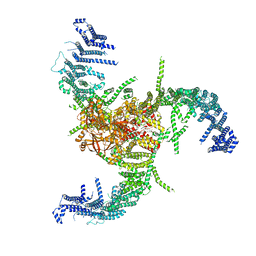 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.81 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7XLT
 
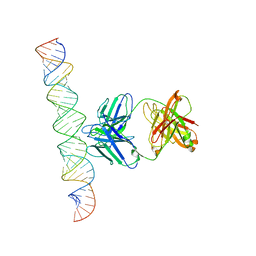 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | Descriptor: | DNA, RNA, S9.6 Fab HC, ... | | Authors: | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
7VLQ
 
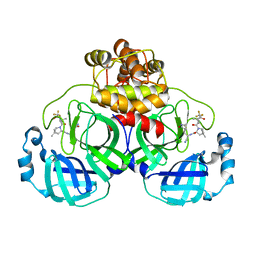 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.939106 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7DR9
 
 | |
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
7YRZ
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, Y.R, Zeng, P, Zhou, X.L, Lin, C, Zhang, J, Yin, X.S, Li, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
6FSE
 
 | | Mus musculus acetylcholinesterase in complex with 1-(4-(4-Ethylpiperazin-1-yl)piperidin-1-yl)-2-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)ethanone dihydrochloride (15) | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]-2-[4-(4-methoxyphenyl)phenoxy]ethanone, 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
6FSD
 
 | | Mus musculus acetylcholinesterase in complex with 2-(4-Biphenylyloxy)-N-[3-(1-piperidinyl)propyl]-acetamide hydrochloride (10) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(4-phenylphenoxy)-~{N}-(3-piperidin-1-ylpropyl)ethanamide, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
4Y0Y
 
 | | Trypsin in complex with with BPTI | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y0Z
 
 | | Trypsin in complex with with BPTI mutant AMINOBUTYRIC ACID | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y11
 
 | | Trypsin in complex with with BPTI mutant (2S)-2-amino-4,4,4-trifluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y10
 
 | | Trypsin in complex with with BPTI mutant (2S)-2-amino-4,4-difluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
