2VGL
 
 | | AP2 CLATHRIN ADAPTOR CORE | | Descriptor: | ADAPTOR PROTEIN COMPLEX AP-2, ALPHA 2 SUBUNIT, AP-2 COMPLEX SUBUNIT BETA-1, ... | | Authors: | Owen, D.J, Collins, B.M, McCoy, A.J, Evans, P.R. | | Deposit date: | 2007-11-14 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture and Functional Model of the Endocytic Ap2 Complex
Cell(Cambridge,Mass.), 109, 2002
|
|
1R4X
 
 | | Crystal Structure Analys of the Gamma-COPI Appendage domain | | Descriptor: | Coatomer gamma subunit, MAGNESIUM ION | | Authors: | Watson, P.J, Frigerio, G, Collins, B.M, Duden, R, Owen, D.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gamma-COP appendage domain - structure and function
Traffic, 5, 2004
|
|
2C3Q
 
 | | Human glutathione-S-transferase T1-1 W234R mutant, complex with S- hexylglutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
4GXB
 
 | | Structure of the SNX17 atypical FERM domain bound to the NPxY motif of P-selectin | | Descriptor: | GLYCEROL, P-selectin, Sorting nexin-17 | | Authors: | Ghai, R, Bugarcic, A, Liu, H, Norwood, S.J, Li, S.S, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2012-09-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Q4G
 
 | | 2.0 Angstrom Crystal Structure of Ovine Prostaglandin H2 Synthase-1, in complex with alpha-methyl-4-biphenylacetic acid | | Descriptor: | 2-(1,1'-BIPHENYL-4-YL)PROPANOIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Gupta, K, Selinksy, B.S, Kaub, C.J, Katz, A.K, Loll, P.J. | | Deposit date: | 2003-08-03 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Crystal Structure of Prostaglandin H(2) Synthase-1: Structural Insights into an Unusual Peroxidase
J.Mol.Biol., 335, 2004
|
|
2AYL
 
 | | 2.0 Angstrom Crystal Structure of Manganese Protoporphyrin IX-reconstituted Ovine Prostaglandin H2 Synthase-1 Complexed With Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, GLYCEROL, ... | | Authors: | Gupta, K, Selinsky, B.S, Loll, P.J. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2V8S
 
 | | VTI1B HABC DOMAIN - EPSINR ENTH DOMAIN COMPLEX | | Descriptor: | CLATHRIN INTERACTOR 1, GLYCEROL, VESICLE TRANSPORT THROUGH INTERACTION WITH T-SNARES HOMOLOG 1B | | Authors: | Owen, D.J, McCoy, A.J, Collins, B.M, Miller, S.E. | | Deposit date: | 2007-08-14 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Snare-Adaptor Interaction is a New Mode of Cargo Recognition in Clathrin Coated Vesicles
Nature, 450, 2007
|
|
1L3P
 
 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2G30
 
 | | beta appendage of AP2 complexed with ARH peptide | | Descriptor: | 16-mer peptide from Low density lipoprotein receptor adapter protein 1, AP-2 complex subunit beta-1, peptide sequence AAF | | Authors: | Edeling, M.A, Collins, B.M, Traub, L.M, Owen, D.J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Switches Involving the AP-2 beta2 Appendage Regulate Endocytic Cargo Selection and Clathrin Coat Assembly
Dev.Cell, 10, 2006
|
|
3PUK
 
 | | Re-refinement of the crystal structure of Munc18-3 and Syntaxin4 N-peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 3 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.054 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PUJ
 
 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PSO
 
 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3RBL
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | CHLORIDE ION, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4I6O
 
 | | Crystal structure of chemically synthesized human anaphylatoxin C3a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Complement C3 | | Authors: | Wang, C.I.A, Ghassemian, A, Collins, B, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-27 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Efficient chemical synthesis of human complement protein C3a.
Chem.Commun.(Camb.), 49, 2013
|
|
3RCH
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the open conformation with LLP and PLP bound to Chain-A and Chain-B respectively | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aromatic L-amino acid decarboxylase | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-31 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RBF
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | Deposit date: | 2011-03-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5OFY
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at pH 9.0. 2.8 Ang; internal aldimine with PLP in the active site | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5UF1
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 in complex with H-trisaccharide | | Descriptor: | CHLORIDE ION, O13, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
1PL2
 
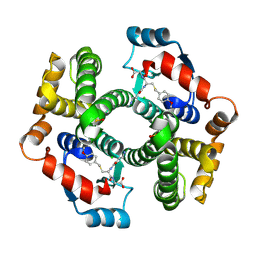 | | Crystal structure of human glutathione transferase (GST) A1-1 T68E mutant in complex with decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5OG0
 
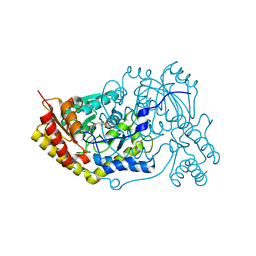 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
3PSN
 
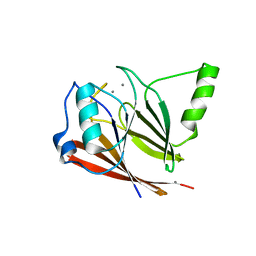 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
1PKW
 
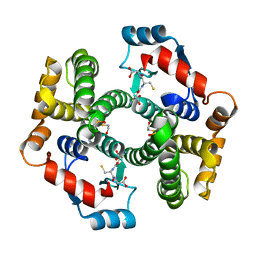 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1QIP
 
 | | HUMAN GLYOXALASE I COMPLEXED WITH S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (LACTOYLGLUTATHIONE LYASE), S-P-NITROBENZYLOXYCARBONYLGLUTATHIONE, ... | | Authors: | Cameron, A.D, Ridderstrom, M, Olin, B, Mannervik, B. | | Deposit date: | 1999-06-14 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue.
Biochemistry, 38, 1999
|
|
