2FK6
 
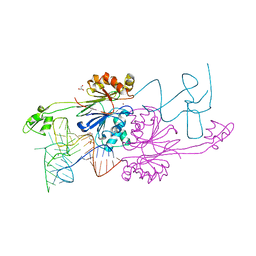 | | Crystal Structure of RNAse Z/tRNA(Thr) complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, RIBONUCLEASE Z, ... | | Authors: | Li de la Sierra-Gallay, I, Mathy, N, Pellegrini, O, Condon, C. | | Deposit date: | 2006-01-04 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5NW7
 
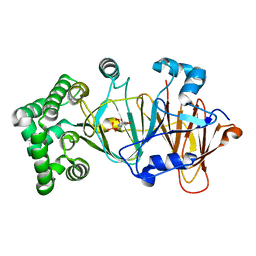 | | Crystal structure of candida albicans phosphomannose isomerase in complex with inhibitor | | Descriptor: | Mannose-6-phosphate isomerase, ZINC ION, [(2~{R},3~{R},4~{S})-5-diazanyl-2,3,4-tris(oxidanyl)-5-oxidanylidene-pentyl] dihydrogen phosphate | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2017-05-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of phosphomannose isomerase from Candida albicans complexed with 5-phospho-d-arabinonhydrazide.
FEBS Lett., 592, 2018
|
|
6HWP
 
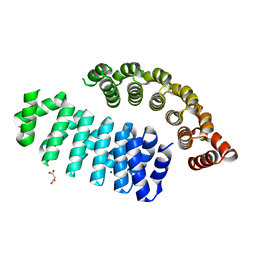 | |
5MTZ
 
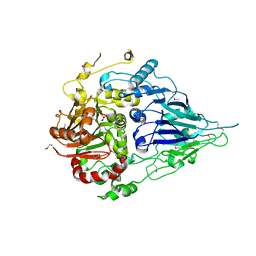 | | Crystal structure of a long form RNase Z from yeast | | Descriptor: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | Authors: | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-06-21 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
3D9W
 
 | | Crystal Structure Analysis of Nocardia farcinica Arylamine N-acetyltransferase | | Descriptor: | Putative acetyltransferase | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F, Martins, M, Dupret, J.M. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural characterization of the arylamine N-acetyltransferase from the opportunistic pathogen Nocardia farcinica
J.Mol.Biol., 383, 2008
|
|
3LNB
 
 | | Crystal Structure Analysis of Arylamine N-acetyltransferase C from Bacillus anthracis | | Descriptor: | COENZYME A, FORMIC ACID, N-acetyltransferase family protein | | Authors: | Li de la Sierra-Gallay, I, Pluvinage, B, Rodrigues-Lima, F. | | Deposit date: | 2010-02-02 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Bacillus anthracis arylamine N-acetyltransferase ((BACAN)NAT1) that inactivates sulfamethoxazole, reveals unusual structural features compared with the other NAT isoenzymes.
Febs Lett., 585, 2011
|
|
1ODF
 
 | | Structure of YGR205w protein. | | Descriptor: | GLYCEROL, HYPOTHETICAL 33.3 KDA PROTEIN IN ADE3-SER2 INTERGENIC REGION, SULFATE ION | | Authors: | Li De La Sierra-Gallay, I, Van Tilbeurgh, H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ygr205W Protein from Saccharomyces Cerevisiae: Close Structural Resemblance to E.Coli Pantothenate Kinase
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
6XUI
 
 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5-PHOSPHOARABINONIC ACID, GLYCEROL, ... | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
6XUH
 
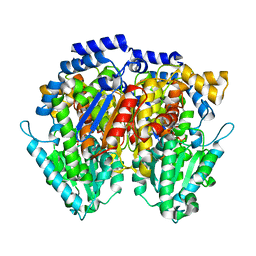 | | Crystal structure of human phosphoglucose isomerase in complex with inhibitor | | Descriptor: | (2R,3R,4S)-5-((2-aminoethyl)amino)-2,3,4-trihydroxy-5-oxopentyl dihydrogen phosphate, 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2020-01-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Novel N-substituted 5-phosphate-d-arabinonamide derivatives as strong inhibitors of phosphoglucose isomerases: Synthesis, structure-activity relationship and crystallographic studies.
Bioorg.Chem., 102, 2020
|
|
6FT5
 
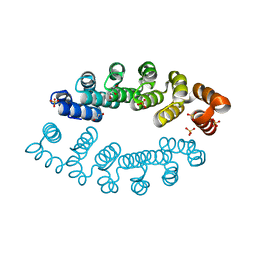 | | Structure of A3_A3, an artificial bi-domain protein based on two identical alphaRep A3 domains | | Descriptor: | GLYCEROL, SULFATE ION, alphaRep A3_A3 | | Authors: | Li de la Sierra-Gallay, I, Leger, C, Di Meo, T. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 9, 2019
|
|
6FSQ
 
 | |
6SX4
 
 | |
6RCY
 
 | |
6SWZ
 
 | |
8AAM
 
 | |
8AB4
 
 | |
8AZG
 
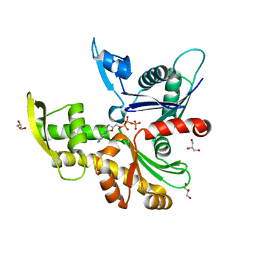 | |
4P78
 
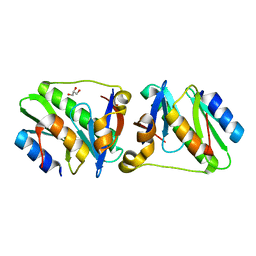 | | HicA3 and HicB3 toxin-antitoxin complex | | Descriptor: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4II9
 
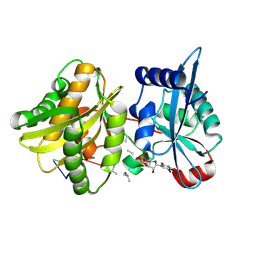 | | Crystal structure of Weissella viridescens FemXVv non-ribosomal amino acid transferase in complex with a peptidyl-RNA conjugate | | Descriptor: | 5-mer peptide, FemX, GLYCEROL, ... | | Authors: | Li de la Sierra-Gallay, I, Fonvielle, M, van Tilbeurgh, H, Arthur, M, Etheve-Quelquejeu, M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of FemXWv in Complex with a Peptidyl-RNA Conjugate: Mechanism of Aminoacyl Transfer from Ala-tRNA(Ala) to Peptidoglycan Precursors
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4P7D
 
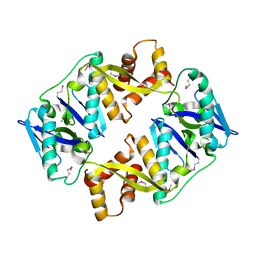 | | Antitoxin HicB3 crystal structure | | Descriptor: | Antitoxin HicB3, CHLORIDE ION | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
8AW4
 
 | |
7Z6K
 
 | |
7Z6A
 
 | |
7Z5Z
 
 | |
7Z5Y
 
 | |
