4M2T
 
 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-SSS | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
5WST
 
 | | Crystal structure of Myo7a SAH | | Descriptor: | Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
7X8U
 
 | |
7FIX
 
 | |
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | Authors: | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C2Z
 
 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
8HHE
 
 | |
5YIQ
 
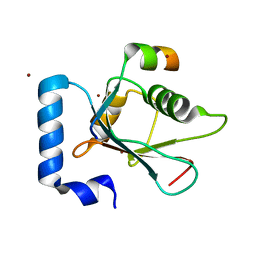 | | Crystal structure of AnkG LIR/LC3B complex | | Descriptor: | Ankyrin-3, Microtubule-associated proteins 1A/1B light chain 3B, ZINC ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIP
 
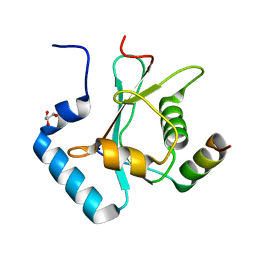 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
7DVM
 
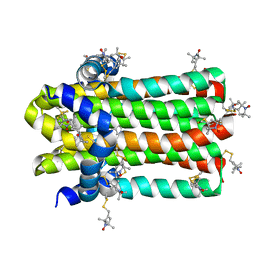 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
7CDB
 
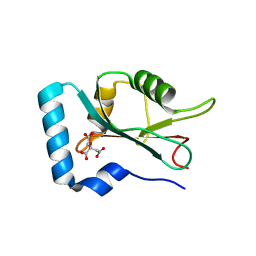 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | Descriptor: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | Authors: | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
7U20
 
 | | Crystal structure of human METTL1 and WDR4 complex | | Descriptor: | SULFATE ION, tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Li, J, Nowak, R.P, Fischer, E.S, Gregory, R. | | Deposit date: | 2022-02-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
7WK3
 
 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7DP3
 
 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
8I54
 
 | | Lb2Cas12a RNA DNA complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3'), Lb2Cas12a, ... | | Authors: | Li, J, Sivaraman, J, Satoru, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
7WI7
 
 | | Crystal structure of human MCM8/9 complex | | Descriptor: | DNA helicase MCM8, DNA helicase MCM9, ZINC ION | | Authors: | Li, J, Liu, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Crystal structure of human MCM8/9 complex
To Be Published
|
|
7E29
 
 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Smolle, M, Liang, H, Liu, Y. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
