8Y38
 
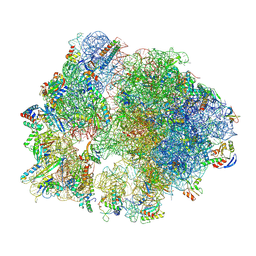 | | Cryo-EM structure of Staphylococcus aureus 70S ribosome (strain 15B196) in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
8Y37
 
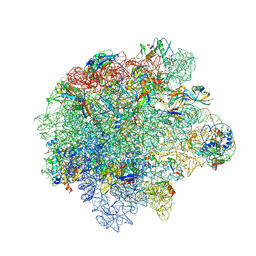 | | Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
2R2N
 
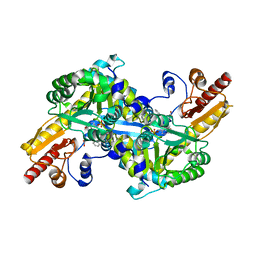 | | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2007-08-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase II.
J.Biol.Chem., 283, 2008
|
|
8Y36
 
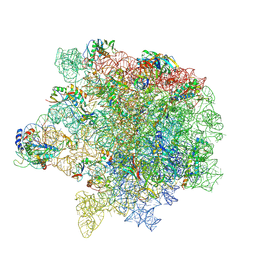 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
2Q8F
 
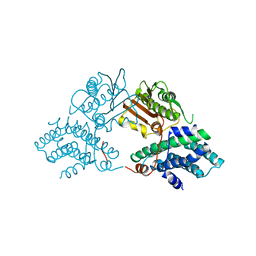 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2QLR
 
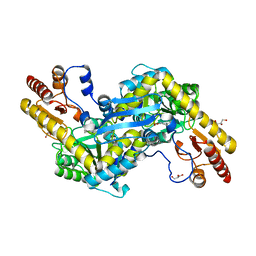 | |
4ELX
 
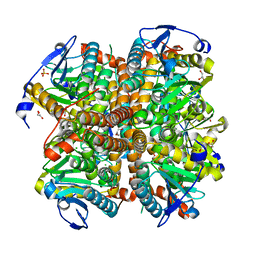 | | Structure of apo E.coli. 1,4-dihydroxy-2- naphthoyl CoA synthases with Cl | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Sun, Y.R, Song, H.G, Li, J, Jiang, M, Li, Y, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-04-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways
Biochemistry, 51, 2012
|
|
1CRW
 
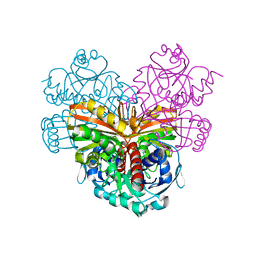 | |
5TPU
 
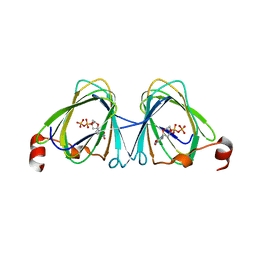 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
4Y07
 
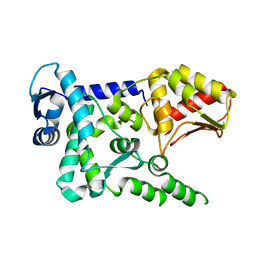 | | Crystal structure of the HECT domain of human WWP2 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Gong, W, Li, J, Li, Z, Xu, Y. | | Deposit date: | 2015-02-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Structure of the HECT domain of human WWP2
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8IGB
 
 | |
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
4Z8Q
 
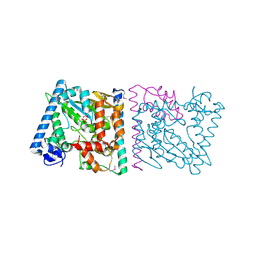 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 COMPLEX, SELENOMETHIONINE SUBSTITUTED. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8U
 
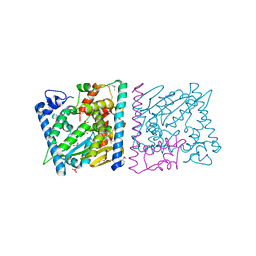 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:-ORF2 WITH ATP | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8T
 
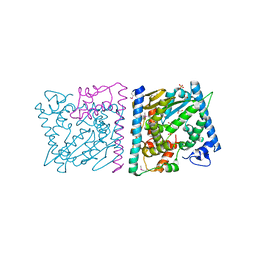 | | CRYSTAL STRUCTURE OF AvrRxo1-ORF1:AvrRxo1-ORF2 WITH SULPHATE IONS | | Descriptor: | ACETATE ION, AvrRxo1-ORF1, AvrRxo1-ORF2, ... | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
4Z8V
 
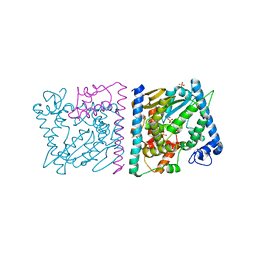 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
2ZKJ
 
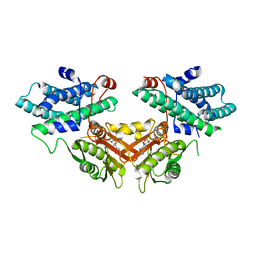 | | Crystal structure of human PDK4-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-03-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity
J.Biol.Chem., 283, 2008
|
|
2QLD
 
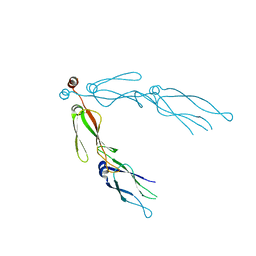 | | human Hsp40 Hdj1 | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Hu, J, Wu, Y, Li, J, Fu, Z, Sha, B. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the putative peptide-binding fragment from the human Hsp40 protein Hdj1.
Bmc Struct.Biol., 8, 2008
|
|
1KTJ
 
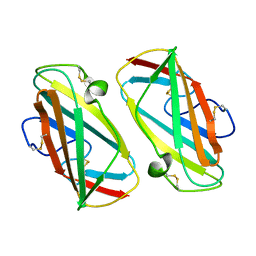 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
2QTW
 
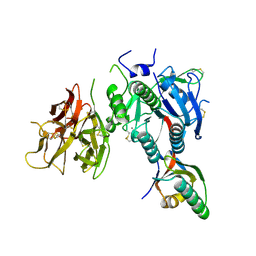 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NX2
 
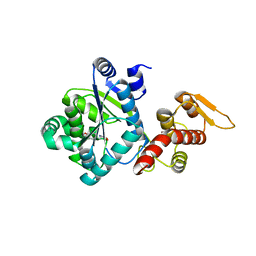 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAI
 
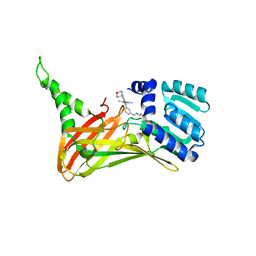 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
6N42
 
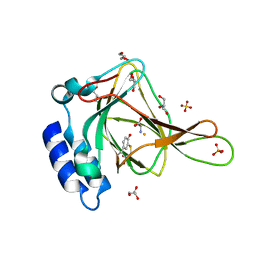 | |
5YZ1
 
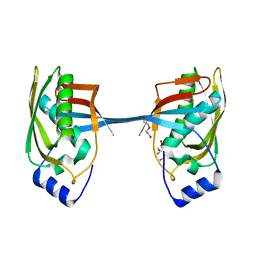 | | Crystal structure of human Archease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein archease | | Authors: | Duan, S.Y, Li, J.X. | | Deposit date: | 2017-12-11 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human archease, a key cofactor of tRNA splicing ligase complex.
Int.J.Biochem.Cell Biol., 122, 2020
|
|
