5YZL
 
 | |
7C2Y
 
 | | The crystal structure of COVID-2019 main protease in the apo state | | 分子名称: | 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-05-10 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | COVID-2019 main protease in the apo state
To Be Published
|
|
7CCD
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | 分子名称: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | 著者 | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | 登録日 | 2020-06-16 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
4I5Q
 
 | | Crystal structure and catalytic mechanism for peroplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium | | 分子名称: | MAGNESIUM ION, Thiol:disulfide interchange protein DsbC | | 著者 | Ha, N.C, Li, J, Kim, J.S, Yoon, B.Y, Yeom, J.H, Lee, K. | | 登録日 | 2012-11-28 | | 公開日 | 2013-10-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | Crystal structure of the periplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium and the mechanistic implications.
J.Struct.Biol., 183, 2013
|
|
4ILF
 
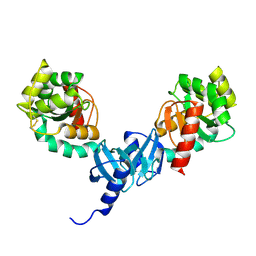 | | Crystal structure of DsbC R125A from Salmonella enterica serovar Typhimurium | | 分子名称: | Thiol:disulfide interchange protein DsbC | | 著者 | Ha, N.C, Li, J, Kim, J.S, Yoon, B.Y, Yeom, J.H, Lee, K. | | 登録日 | 2012-12-31 | | 公開日 | 2013-10-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Crystal structure of the periplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium and the mechanistic implications.
J.Struct.Biol., 183, 2013
|
|
7CBY
 
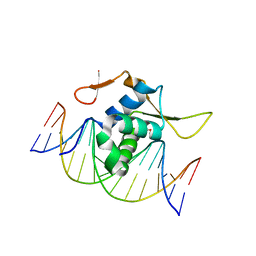 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | 著者 | Dai, S.Y, Li, J, Chen, Y.H. | | 登録日 | 2020-06-15 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.646 Å) | | 主引用文献 | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
7F42
 
 | |
7F41
 
 | |
7F43
 
 | |
7DR8
 
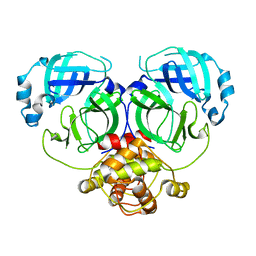 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-26 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.338149 Å) | | 主引用文献 | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7DR9
 
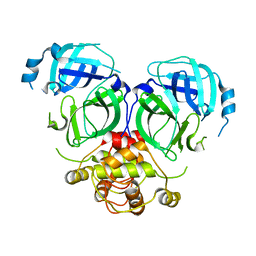 | |
7DRA
 
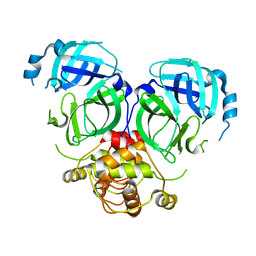 | |
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | 分子名称: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J, Zhu, R, Pei, Y. | | 登録日 | 2021-03-23 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-04-06 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-03-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F4G
 
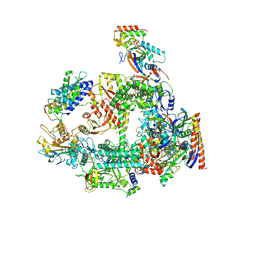 | | Structure of RPAP2-bound RNA polymerase II | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-06-18 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
4H7Q
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase in complex with alpha-ketoisocaproic acid and ADP | | 分子名称: | 2-OXO-4-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H85
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-alpha-chloroisocaproate complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALPHA-CHLOROISOCAPROIC ACID, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H81
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-2-chloro-3-phenylpropanoic acid complex with ADP | | 分子名称: | (2R)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7C5F
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 1.88 Angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Zhang, L, Liu, M.R, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | 登録日 | 2020-05-20 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Characterization and structure of glyceraldehyde-3-phosphate dehydrogenase type 1 from Escherichia coli.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7ENJ
 
 | | Human Mediator (deletion of MED1-IDR) in a Tail-bent conformation (MED-B) | | 分子名称: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, ... | | 著者 | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | 登録日 | 2021-04-17 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | 登録日 | 2021-04-16 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (4.07 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | 登録日 | 2021-04-16 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (4.13 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7EMF
 
 | | Human Mediator (deletion of MED1-IDR) in a Tail-extended conformation | | 分子名称: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 16, Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | 登録日 | 2021-04-13 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
3TZ5
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex with ADP | | 分子名称: | 4-PHENYL-BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-27 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
