5X3V
 
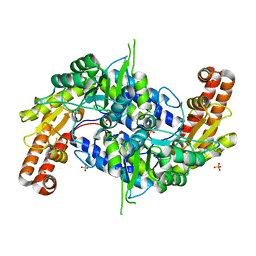 | | Structure of human SHMT2 protein mutant | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase, mitochondrial | | Authors: | Li, J, Sheng, J. | | Deposit date: | 2017-02-09 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of human SHMT2 protein mutant
To Be Published
|
|
8IDC
 
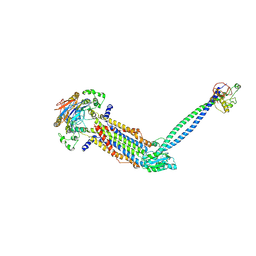 | |
8IGQ
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
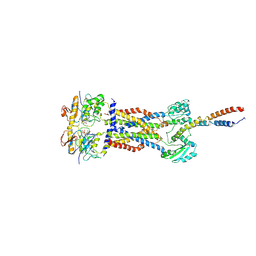
8IDD
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
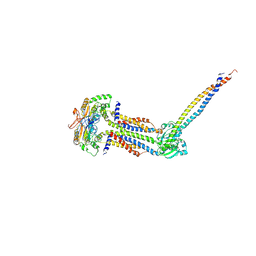
8IDB
 
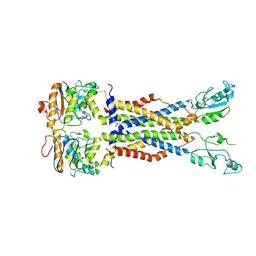 | |
8JIA
 
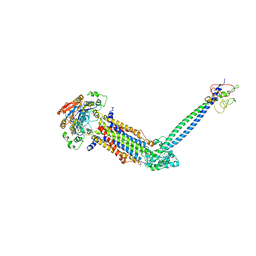 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-05-26 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
6BLK
 
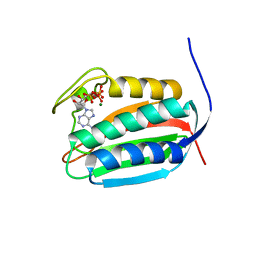 | | Mycobacterial sensor histidine kinase MprB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal transduction histidine-protein kinase/phosphatase mprB | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Mycobacterial sensor histidine kinase MprB
to be published
|
|
4PY4
 
 | |
5XR9
 
 | | Crystal structure of Human Hsp90 with FS6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-bromanyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Human Hsp90 with FS6
To Be Published
|
|
5XQD
 
 | | Crystal structure of Human Hsp90 with FS2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS2
To Be Published
|
|
5XR5
 
 | | Crystal structure of Human Hsp90 with FS4 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2,2-dimethyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human Hsp90 with FS4
To Be Published
|
|
5XRB
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5
To Be Published
|
|
2JQW
 
 | |
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
5XRE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(4-methoxyphenyl)-3-[5-(8-methylquinolin-5-yl)-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1
To Be Published
|
|
4M1M
 
 | |
7VEZ
 
 | |
7VF0
 
 | |
5XBF
 
 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSJ
 
 | | XylFII-LytSN complex | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSD
 
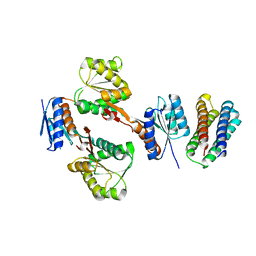 | | XylFII-LytSN complex mutant - D103A | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WK2
 
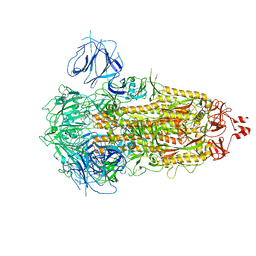 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVO
 
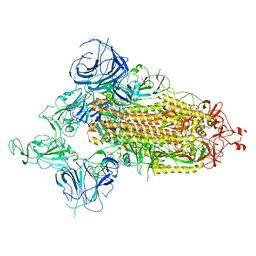 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
