7DW5
 
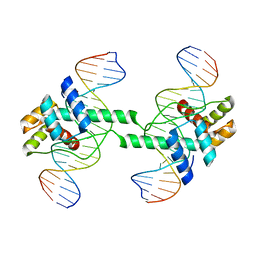 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | Descriptor: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | Authors: | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
7KCB
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NAD+ and Trifluoroethanol | | Descriptor: | ADH1 isoform 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KCQ
 
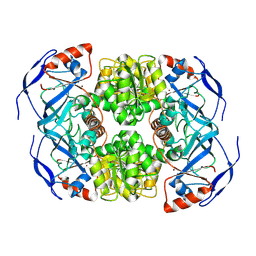 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Open Form of Apoenzyme | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
7KC2
 
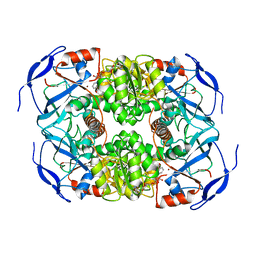 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
3MGM
 
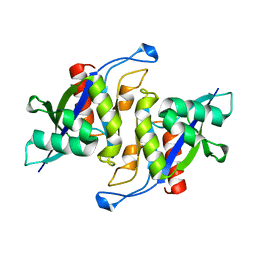 | | Crystal structure of human NUDT16 | | Descriptor: | U8 snoRNA-decapping enzyme | | Authors: | Yan, J, Lu, G, Zhang, J, Qi, J, Li, Z, Gao, F. | | Deposit date: | 2010-04-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure and the template mRNA decapping activity of human NUDT16
To be Published
|
|
7JZZ
 
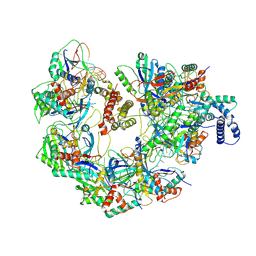 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF14 | | Descriptor: | AcrF14, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
3MCN
 
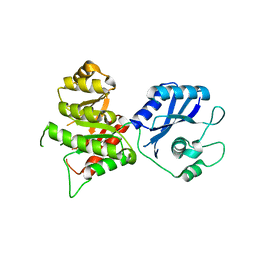 | | Crystal Structure of the 6-hyroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis | | Descriptor: | 2,6-diamino-5-nitropyrimidin-4(3H)-one, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, MAGNESIUM ION | | Authors: | Pemble IV, C.W, Mehta, P.K, Mehra, S, Li, Z, Lee, R.E, White, S.W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
Plos One, 5, 2010
|
|
7JZX
 
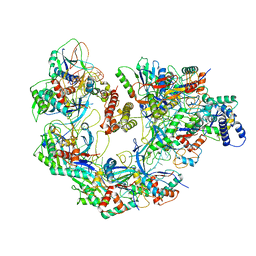 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF7 | | Descriptor: | AcrF7, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZW
 
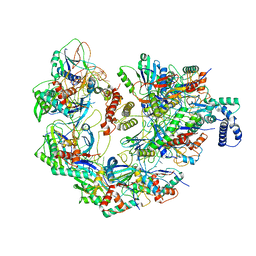 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF4 | | Descriptor: | CRISPR repeat sequence, CRISPR type I-F/YPEST-associated protein Csy1, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
3MCM
 
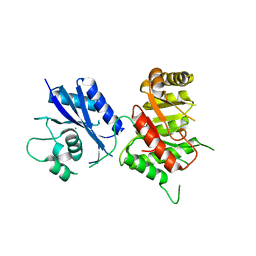 | | Crystal Structure of the 6-hyroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, MAGNESIUM ION | | Authors: | Pemble IV, C.W, Mehta, P.K, Mehra, S, Li, Z, Lee, R.E, White, S.W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
Plos One, 5, 2010
|
|
3MCO
 
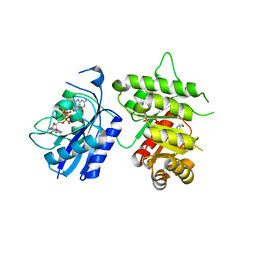 | | Crystal Structure of the 6-hyroxymethyl-7,8-dihydropterin pyrophosphokinase dihydropteroate synthase bifunctional enzyme from Francisella tularensis | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Pemble IV, C.W, Mehta, P.K, Mehra, S, Li, Z, Lee, R.E, White, S.W. | | Deposit date: | 2010-03-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase.dihydropteroate synthase bifunctional enzyme from Francisella tularensis.
Plos One, 5, 2010
|
|
3MHD
 
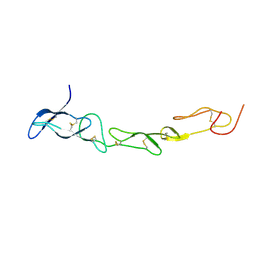 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3RKZ
 
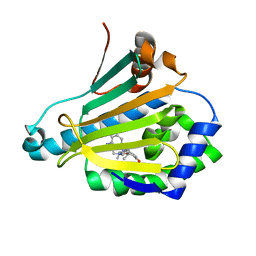 | | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. | | Descriptor: | (5R,6S)-3-(L-alanyl)-5,6,15,15,18-pentamethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,5,9]triazacyclotetradecino[1,2-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, Li, Z, Dushin, R.G, Nittoli, T, Otteng, M, Nikitenko, A, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Olland, A, Johnson, M, Levin, J.I. | | Deposit date: | 2011-04-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5693 Å) | | Cite: | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor which significantly decreases tumor volume in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7JZY
 
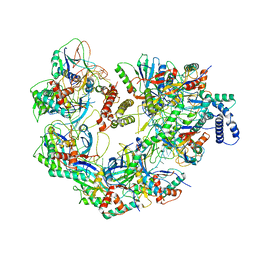 | | CryoEM structure of a CRISPR-Cas complex | | Descriptor: | AcrF9, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of a CRISPR-Cas complex
To Be Published
|
|
7L48
 
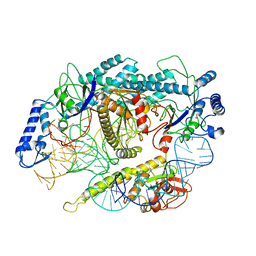 | | Cryo-EM structure of a CRISPR-Cas12f Binary Complex | | Descriptor: | Cas12f, ZINC ION, sgRNA | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
7L49
 
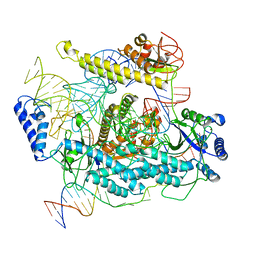 | | Cryo-EM structure of CRISPR-Cas12f Ternary Complex | | Descriptor: | Cas12f1, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
3M9I
 
 | |
3MI8
 
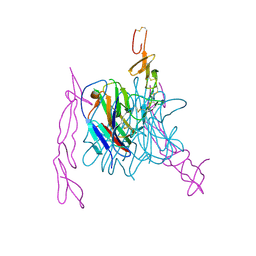 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3OV9
 
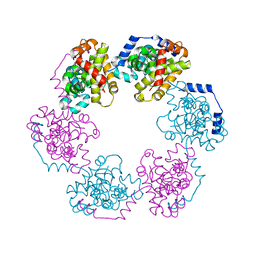 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
7KJY
 
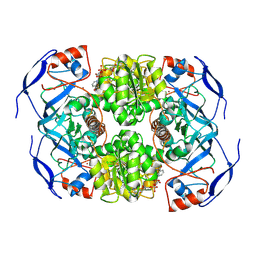 | | Symmetry in Yeast Alcohol Dehydrogenase 1 - Open Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V, Guntupalli, S.R. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
3SHB
 
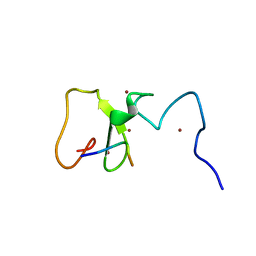 | | Crystal Structure of PHD Domain of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 peptide, ZINC ION | | Authors: | Hu, L, Li, Z, Wang, P, Lin, Y, Xu, Y. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2.
Cell Res., 2011
|
|
3OUO
 
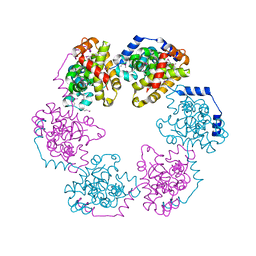 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-05-25 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
7CIP
 
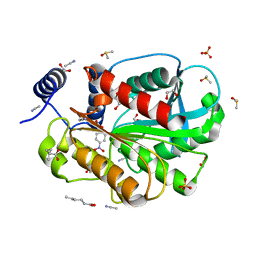 | | Microbial Hormone-sensitive lipase E53 wild type | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 wild type
To Be Published
|
|
5XWP
 
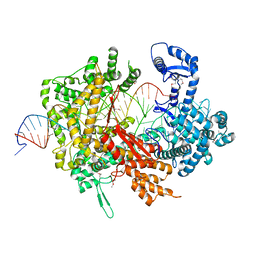 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
8FAR
 
 | |
