2LV4
 
 | | ZirS C-terminal Domain | | Descriptor: | Putative outer membrane or exported protein | | Authors: | Prehna, G, Li, Y, Stoynov, N, Okon, M, Vukovic, M, Mcintosh, L.P, Foster, L.J, Finlay, B, Strynadka, N.C.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The zinc regulated antivirulence pathway of salmonella is a multiprotein immunoglobulin adhesion system.
J.Biol.Chem., 287, 2012
|
|
2M3E
 
 | |
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
3F0N
 
 | | Mus Musculus Mevalonate Pyrophosphate Decarboxylase | | Descriptor: | MEVALONATE PYROPHOSPHATE DECARBOXYLASE, PHOSPHATE ION | | Authors: | Walker, J.R, Davis, T, Vesterberg, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Mus Musculus Mevalonate Pyrophosphate Decarboxylase
To be Published
|
|
5TEY
 
 | | Human METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-23 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human METTL3-METTL14 complex
to be published
|
|
3CLZ
 
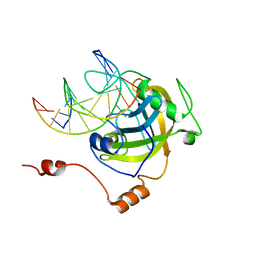 | | The set and ring associated (SRA) domain of UHRF1 bound to methylated DNA | | Descriptor: | 5'-D(*DCP*DCP*DCP*DTP*DGP*DCP*DGP*DGP*DGP*DCP*DCP*DC)-3', 5'-D(*DGP*DGP*DGP*DCP*DCP*(5CM)P*DGP*DCP*DAP*DGP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
3CWD
 
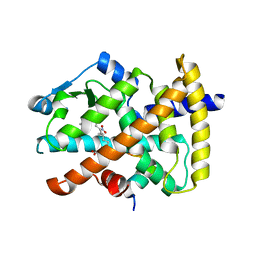 | | Molecular recognition of nitro-fatty acids by PPAR gamma | | Descriptor: | (9E,12Z)-10-nitrooctadeca-9,12-dienoic acid, (9Z,12E)-12-nitrooctadeca-9,12-dienoic acid, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Martynowski, D, Li, Y. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of nitrated fatty acids by PPAR gamma.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2MQ9
 
 | |
4ILD
 
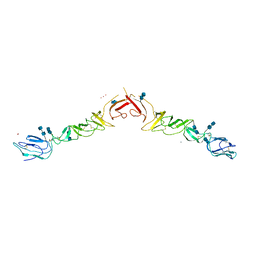 | | Crystal structure of truncated Bovine viral diarrhea virus 1 E2 envelope protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein E2, ... | | Authors: | Modis, Y, Li, Y, Wang, J. | | Deposit date: | 2012-12-30 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structure of glycoprotein E2 from bovine viral diarrhea virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3E19
 
 | | Crystal Structure of Iron Uptake Regulatory Protein (FeoA) Solved by Sulfur SAD in a Monoclinic Space Group | | Descriptor: | FeoA, GLYCEROL, PHOSPHATE ION | | Authors: | Hughes, R.C, Li, Y, Wang, B.-C, Liu, Z.-J, Ng, J.D. | | Deposit date: | 2008-08-02 | | Release date: | 2008-12-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Structure Determination of Iron Uptake Regulatory
Protein (FeoA) by Sulfur SAD in a Monoclinic Space Group
To be Published
|
|
4MVT
 
 | | Crystal structure of SUMO E3 Ligase PIAS3 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase PIAS3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SUMO E3 Ligase PIAS3
to be published
|
|
3DKM
 
 | | Crystal structure of the HECTD1 CPH domain | | Descriptor: | E3 ubiquitin-protein ligase HECTD1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the CPH domain of the E3 ubiquitin-protein ligase HECTD1.
To be Published
|
|
3G1N
 
 | | Catalytic domain of the human E3 ubiquitin-protein ligase HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, SODIUM ION | | Authors: | Walker, J.R, Qiu, L, Li, Y, Davis, T, Tempel, W, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hect Domain of Human HUWE1/MULE
To be Published
|
|
2P2O
 
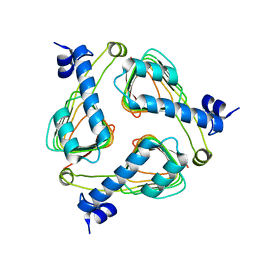 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
4JUY
 
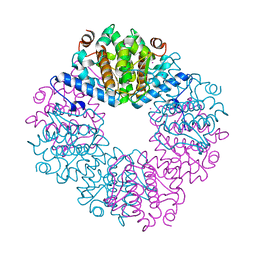 | | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31 | | Descriptor: | E3 ubiquitin-protein ligase RNF31, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31
To be Published
|
|
2PG0
 
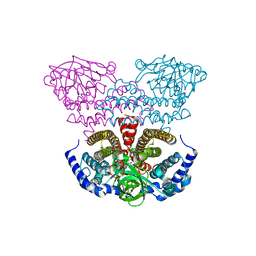 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
2P91
 
 | | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chen, L, Li, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5
To be Published
|
|
3FCX
 
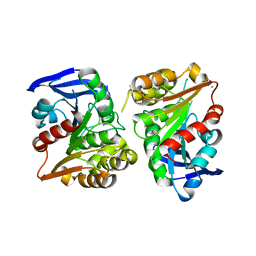 | | Crystal structure of human esterase D | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-formylglutathione hydrolase | | Authors: | Wu, D, Li, Y, Song, G, Zhang, D, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human esterase D: a potential genetic marker of retinoblastoma
Faseb J., 23, 2009
|
|
3FL2
 
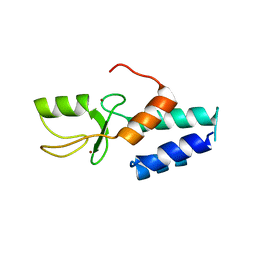 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
4O2W
 
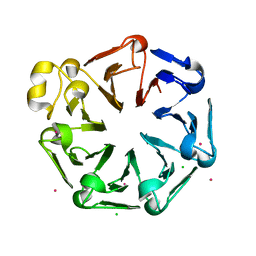 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
3GUC
 
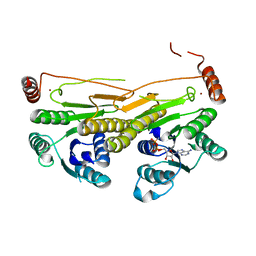 | | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ubiquitin-like modifier-activating enzyme 5, ZINC ION | | Authors: | Walker, J.R, Bacik, J.P, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human Ubiquitin-activating Enzyme 5 in Complex with AMPPNP
To be Published
|
|
2MQ6
 
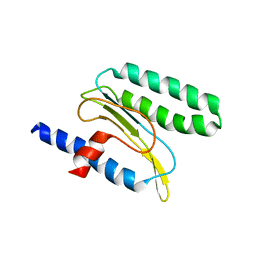 | |
1WM8
 
 | | Solution Structure of BmP03 from the Venom of Scorpion Buthus martensii Karsch, 10 structures | | Descriptor: | Neurotoxin BmP03 | | Authors: | Wu, H, He, F, Li, Y, Wu, G, Cao, C, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of BmP03 from Venom of Scorpion Buthus martensii Karsch
Acta Chim.Sinica, 58, 2000
|
|
1WT8
 
 | | Solution Structure of BmP08 from the Venom of Scorpion Buthus martensii Karsch, 20 structures | | Descriptor: | Neurotoxin BmK X | | Authors: | Wu, H, Chen, X, Tong, X, Li, Y, Zhang, N, Wu, G. | | Deposit date: | 2004-11-17 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP08, a novel short-chain scorpion toxin from Buthus martensi Karsch.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
1SBZ
 
 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
