4JYY
 
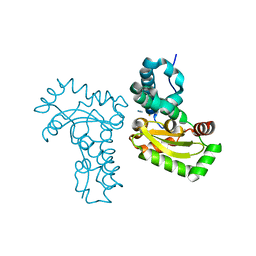 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | Descriptor: | AZIDE ION, FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
4JZG
 
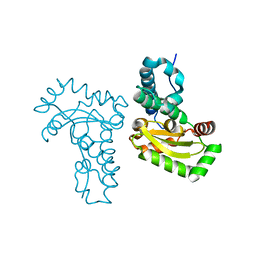 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
6KMH
 
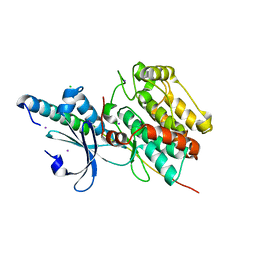 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
7W30
 
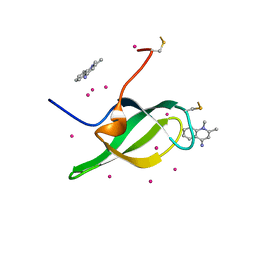 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
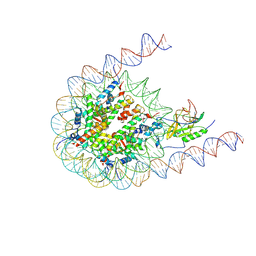 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
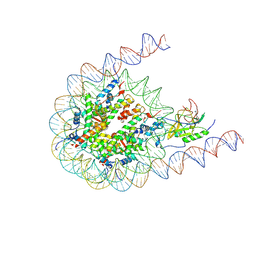 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
6JCI
 
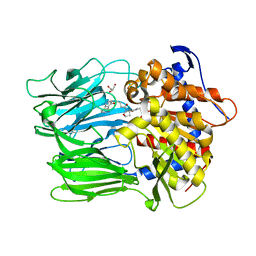 | |
7W2P
 
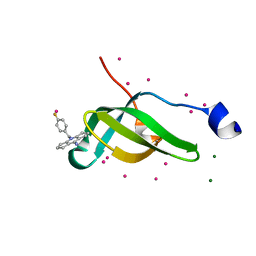 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 2-[(4-fluorophenyl)methyl]-2-azatricyclo[7.3.0.0^{3,7}]dodeca-1(9),3(7)-dien-8-imine, MAGNESIUM ION, Survival motor neuron protein, ... | | Authors: | Li, W, Arrowsmith, C.H, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7CY6
 
 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY7
 
 | | Crystal Structure of CMD1 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY8
 
 | | Crystal Structure of CMD1 in complex with 5mC-DNA and vitamin C | | Descriptor: | 1,2-ETHANEDIOL, ASCORBIC ACID, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY4
 
 | | Crystal Structure of CMD1 in apo form | | Descriptor: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY5
 
 | | Crystal Structure of CMD1 in complex with vitamin C | | Descriptor: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
3KBH
 
 | | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Wu, K, Li, W, Peng, G, Li, F. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
5OWD
 
 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-hept-5-en-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
5OW7
 
 | | VDR complex | | Descriptor: | (3~{S})-3-[(1~{S},3~{a}~{S},4~{E},7~{a}~{S})-7~{a}-methyl-4-[(2~{Z})-2-[(5~{S})-2-methylidene-5-oxidanyl-cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-3-oxidanyl-~{N}-propan-2-yl-butanamide, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
5OW9
 
 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-heptan-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJH
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
